

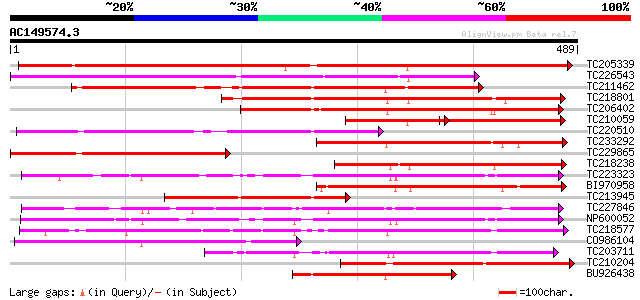
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149574.3 + phase: 0
(489 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC205339 weakly similar to UP|Q6QDB6 (Q6QDB6) UDP-glucose glucos... 558 e-159
TC226543 weakly similar to UP|Q6VAA9 (Q6VAA9) UDP-glycosyltransf... 322 2e-88
TC211462 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltran... 289 2e-78
TC218801 similar to UP|Q8W3P8 (Q8W3P8) ABA-glucosyltransferase, ... 278 5e-75
TC206402 similar to UP|Q8S995 (Q8S995) Glucosyltransferase-14, p... 258 4e-69
TC210059 similar to UP|Q8S9A1 (Q8S9A1) Glucosyltransferase-8 (Fr... 145 1e-67
TC220510 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltran... 239 2e-63
TC233292 similar to UP|Q8S9A0 (Q8S9A0) Glucosyltransferase-9, pa... 234 9e-62
TC229865 weakly similar to UP|P93789 (P93789) UDP-glucose glucos... 211 5e-55
TC218238 weakly similar to UP|Q8H0F2 (Q8H0F2) Anthocyanin 3'-glu... 206 1e-53
TC223323 similar to UP|Q8S3B8 (Q8S3B8) Zeatin O-glucosyltransfer... 193 2e-49
BI970958 193 2e-49
TC213945 similar to UP|Q6QDB6 (Q6QDB6) UDP-glucose glucosyltrans... 192 2e-49
TC227846 weakly similar to UP|Q947K4 (Q947K4) Thiohydroximate S-... 191 6e-49
NP600052 zeatin O-glucosyltransferase [Glycine max] 189 3e-48
TC218577 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (F... 185 3e-47
CO986104 177 7e-45
TC203711 similar to UP|Q8S9A4 (Q8S9A4) Glucosyltransferase-5, pa... 177 9e-45
TC210204 similar to UP|Q9SY84 (Q9SY84) F14N23.30, partial (21%) 174 6e-44
BU926438 similar to PIR|T03747|T03 glucosyltransferase IS5a (EC ... 172 2e-43
>TC205339 weakly similar to UP|Q6QDB6 (Q6QDB6) UDP-glucose
glucosyltransferase, partial (55%)
Length = 1537
Score = 558 bits (1439), Expect = e-159
Identities = 278/484 (57%), Positives = 354/484 (72%), Gaps = 6/484 (1%)
Frame = +2
Query: 8 LHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNIDIQ 67
LHI++FPF G GH IP DMA+ F +GVR TIVTTPLN I + K +I+I
Sbjct: 29 LHIMLFPFPGQGHLIPMSDMARAFNGRGVRTTIVTTPLNVATIRGTI--GKETETDIEIL 202
Query: 68 TIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPHCVVADMFFP 127
T+KFP EAGLPEGCEN +SIPS V F AIR+L+ P E LLLQ +PHC++A FFP
Sbjct: 203 TVKFPSAEAGLPEGCENTESIPSPDLVLTFLKAIRMLEAPLEHLLLQHRPHCLIASAFFP 382
Query: 128 WATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPGNIKMTRL 187
WA+ SA K IPR+VFHGT F+LCAS+C++ YQP+KNVSSDTD F I LPG+I+MTRL
Sbjct: 383 WASHSATKLKIPRLVFHGTGVFALCASECVRLYQPHKNVSSDTDPFIIPHLPGDIQMTRL 562
Query: 188 QLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYY----REVLGIK 243
LP+ + ++ +EIK+SE+ SYG+IVNSFYELE VYADYY +V G +
Sbjct: 563 LLPDYAKTDGDGETGLTRVLQEIKESELASYGMIVNSFYELEQVYADYYDKQLLQVQGRR 742
Query: 244 EWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNSQ 303
W+IGP S+ N++K + RGK+AS+D+ + LKWLD+K NSVVY+CFGS+ +F +Q
Sbjct: 743 AWYIGPLSLCNQDKGK-----RGKQASVDQGDILKWLDSKKANSVVYVCFGSIANFSETQ 907
Query: 304 LKEIAMGLEASGHNFIWVVRTQTEDGDEWLPEGFEERT--EGKGLIIRGWSPQVMILEHE 361
L+EIA GLE SG FIWVVR +D WLPEGFE RT EG+G+II GW+PQV+IL+H+
Sbjct: 908 LREIARGLEDSGQQFIWVVRRSDKDDKGWLPEGFETRTTSEGRGVIIWGWAPQVLILDHQ 1087
Query: 362 AIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKWVMKVG 421
A+GAFVTHCGWNS LE V AGVPM+TWPV+AEQFYNEK VT++L+ GVPVGVKKW VG
Sbjct: 1088AVGAFVTHCGWNSTLEAVSAGVPMLTWPVSAEQFYNEKFVTDILQIGVPVGVKKWNRIVG 1267
Query: 422 DNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALIEELRSLS 481
DN+ +A++KA+ R+M GEEA MRN+A LA+MA A++ +GSSY LI+ LRS++
Sbjct: 1268DNITSNALQKALHRIMIGEEAEPMRNRAHKLAQMATTALQHNGSSYCHFTHLIQHLRSIA 1447
Query: 482 HHQH 485
Q+
Sbjct: 1448SLQN 1459
>TC226543 weakly similar to UP|Q6VAA9 (Q6VAA9) UDP-glycosyltransferase 73E1,
partial (21%)
Length = 1237
Score = 322 bits (826), Expect = 2e-88
Identities = 171/409 (41%), Positives = 246/409 (59%), Gaps = 4/409 (0%)
Frame = +2
Query: 1 MDSQSNPLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIH 60
M+ + L + PF+ H IP +DMA+LFA V VTI+TT N K+++
Sbjct: 26 MEKKKGELKSIFLPFLSTSHIIPLVDMARLFALHDVDVTIITTAHNATVFQKSIDLDASR 205
Query: 61 FNNIDIQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPHCV 120
I + FP + GLP G E + P + + LLQQ FE+L +P +
Sbjct: 206 GRPIRTHVVNFPAAQVGLPVGIEAFNVDTPREMTPRIYMGLSLLQQVFEKLFHDLQPDFI 385
Query: 121 VADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPG 180
V DMF PW+ D+AAK GIPRI+FHG S+ + A+ +++Y P+ DTD F + LP
Sbjct: 386 VTDMFHPWSVDAAAKLGIPRIMFHGASYLARSAAHSVEQYAPHLEAKFDTDKFVLPGLPD 565
Query: 181 NIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVL 240
N++MTRLQLP+ L + +L IK SE +SYG + NSFY+LE+ Y ++Y+ ++
Sbjct: 566 NLEMTRLQLPDWLRS----PNQYTELMRTIKQSEKKSYGSLFNSFYDLESAYYEHYKSIM 733
Query: 241 GIKEWHIGPFSI-HNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHF 299
G K W IGP S+ N++ +++ KE +K LKWL++K +SV+Y+ FGS+ F
Sbjct: 734 GTKSWGIGPVSLWANQDAQDKAARGYAKEEE-EKEGWLKWLNSKAESSVLYVSFGSINKF 910
Query: 300 LNSQLKEIAMGLEASGHNFIWVVR-TQTEDGDEWLPEGFEERTE--GKGLIIRGWSPQVM 356
SQL EIA LE SGH+FIWVVR +GD +L E FE+R + KG +I GW+PQ++
Sbjct: 911 PYSQLVEIARALEDSGHDFIWVVRKNDGGEGDNFLEE-FEKRMKESNKGYLIWGWAPQLL 1087
Query: 357 ILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVL 405
ILE+ AIG V+HCGWN+V+E V + +PM TWP+ AE F+N+ L +L
Sbjct: 1088ILENPAIGGLVSHCGWNTVVESVNSVLPMATWPLFAEHFFNDNLAVYLL 1234
>TC211462 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltransferase,
partial (64%)
Length = 1066
Score = 289 bits (740), Expect = 2e-78
Identities = 155/364 (42%), Positives = 221/364 (60%), Gaps = 9/364 (2%)
Frame = +3
Query: 54 LEQSKIHFNNIDIQTIKFPCVEAGLPEGCENVDSIPSVS-FVPAFFAAIRLLQQPFEELL 112
L QSK N + T FP E GLP+G EN+ ++ + + AA LL++P E +
Sbjct: 9 LHQSK----NFRVHTFDFPSEEVGLPDGVENLSAVTDLEKSYRIYIAATTLLREPIESFV 176
Query: 113 LQQKPHCVVADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDL 172
+ P C+VAD + W D A K IP + F+G S FS+CA + +KK++
Sbjct: 177 ERDPPDCIVADFLYCWVEDLAKKLRIPWLDFNGFSLFSICAMESVKKHR------IGDGP 338
Query: 173 FEITDLPGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENV- 231
F I D P ++ T+ P + + E + + ++S G I+N+F EL+
Sbjct: 339 FVIPDFPDHV---------TIKSTPP--KDMREFLEPLLTAALKSNGFIINNFAELDGEE 485
Query: 232 YADYYREVLGIKEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYM 291
Y +Y + G K WH+GP S+ R + E+ + RG+++ + HECL WLD+K +NSVVY+
Sbjct: 486 YLRHYEKTTGHKAWHLGPASLVRRTEMEK--AERGQKSVVSTHECLSWLDSKRVNSVVYV 659
Query: 292 CFGSMTHFLNSQLKEIAMGLEASGHNFIWVV-------RTQTEDGDEWLPEGFEERTEGK 344
FGS+ +F + QL EIA G+EASG+ FIWVV E+ ++WLP+GFEER K
Sbjct: 660 SFGSLCYFPDKQLYEIACGMEASGYEFIWVVPEKKGKEEESEEEKEKWLPKGFEERK--K 833
Query: 345 GLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEV 404
G+II+GW+PQV+ILEH A+GAF+THCGWNS +E V AGVPMITWPV ++QFYNEKL+T+V
Sbjct: 834 GMIIKGWAPQVVILEHPAVGAFLTHCGWNSTVEAVSAGVPMITWPVHSDQFYNEKLITQV 1013
Query: 405 LKTG 408
G
Sbjct: 1014RGIG 1025
>TC218801 similar to UP|Q8W3P8 (Q8W3P8) ABA-glucosyltransferase, partial
(60%)
Length = 1281
Score = 278 bits (710), Expect = 5e-75
Identities = 155/314 (49%), Positives = 211/314 (66%), Gaps = 17/314 (5%)
Frame = +1
Query: 183 KMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVLGI 242
+MTR QLP L ++ ++ + ++ E +S+G VNSF++LE YA+ + G
Sbjct: 1 EMTRSQLPVFL-------RTPSQFPDRVRQLEEKSFGTFVNSFHDLEPAYAEQVKNKWGK 159
Query: 243 KEWHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNS 302
K W IGP S+ NR E++ + RGK +ID+ +CL WL++K NSV+Y+ FGS+ +
Sbjct: 160 KAWIIGPVSLCNRTAEDK--TERGKLPTIDEEKCLNWLNSKKPNSVLYVSFGSLLRLPSE 333
Query: 303 QLKEIAMGLEASGHNFIWVVRT-------QTEDGD-EWLPEGFEERTE--GKGLIIRGWS 352
QLKEIA GLEAS +FIWVVR E+G+ +LPEGFE+R + KGL++RGW+
Sbjct: 334 QLKEIACGLEASEQSFIWVVRNIHNNPSENKENGNGNFLPEGFEQRMKEKDKGLVLRGWA 513
Query: 353 PQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVG 412
PQ++ILEH AI F+THCGWNS LE V AGVPMITWP++AEQF NEKL+TEVLK GV VG
Sbjct: 514 PQLLILEHVAIKGFMTHCGWNSTLESVCAGVPMITWPLSAEQFSNEKLITEVLKIGVQVG 693
Query: 413 VKKWVMKVGDNVEW------DAVEKAVKRVM-EGEEAYEMRNKAKMLAEMAKKAVEEDGS 465
++W + N EW + VE AV+++M E EEA EM + K +AE AK+AVEE G+
Sbjct: 694 SREW---LSWNSEWKDLVGREKVESAVRKLMVESEEAEEMTTRVKDIAEKAKRAVEEGGT 864
Query: 466 SYSQLNALIEELRS 479
SY+ ALIEEL++
Sbjct: 865 SYADAEALIEELKA 906
>TC206402 similar to UP|Q8S995 (Q8S995) Glucosyltransferase-14, partial (59%)
Length = 1152
Score = 258 bits (659), Expect = 4e-69
Identities = 134/287 (46%), Positives = 198/287 (68%), Gaps = 9/287 (3%)
Frame = +2
Query: 200 SQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVLGIKEWHIGPFSIHNRNKEE 259
++S+ ++ EEI+++E+ SYGV++NSF ELE YA Y+++ G K W IGP S+ N++ +
Sbjct: 2 NESWKQINEEIREAEMSSYGVVMNSFEELEPAYATGYKKIRGDKLWCIGPVSLINKDHLD 181
Query: 260 EIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNSQLKEIAMGLEASGHNFI 319
+ + RG ASID + +KWLD + +V+Y C GS+ + QLKE+ + LEAS FI
Sbjct: 182 K--AQRGT-ASIDVSQHIKWLDCQKPGTVIYACLGSLCNLTTPQLKELGLALEASKRPFI 352
Query: 320 WVVRT--QTEDGDEWLPE-GFEERTEGKGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVL 376
WV+R +E+ ++W+ E GFEERT + L+IRGW+PQ++IL H AIG F+THCGWNS L
Sbjct: 353 WVIREGGHSEELEKWIKEYGFEERTNARSLLIRGWAPQILILSHPAIGGFITHCGWNSTL 532
Query: 377 EGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVK---KW--VMKVGDNVEWDAVEK 431
E + AGVPM+TWP+ A+QF NE LV VLK GV VGV+ W +++G V+ VE+
Sbjct: 533 EAICAGVPMLTWPLFADQFLNESLVVHVLKVGVKVGVEIPLTWGKEVEIGVQVKKKDVER 712
Query: 432 AVKRVM-EGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALIEEL 477
A+ ++M E E+ + R + + LAEMA +AVE+ GSSYS + LI+++
Sbjct: 713 AIAKLMDETSESEKRRKRVRELAEMANRAVEKGGSSYSNVTLLIQDI 853
>TC210059 similar to UP|Q8S9A1 (Q8S9A1) Glucosyltransferase-8 (Fragment),
partial (37%)
Length = 803
Score = 145 bits (365), Expect(2) = 1e-67
Identities = 67/92 (72%), Positives = 78/92 (83%), Gaps = 2/92 (2%)
Frame = +1
Query: 290 YMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDEWLPEGFEERT--EGKGLI 347
Y+CFG MT F ++QLKEIA+GLEASG NFIWVV+ + EWLPEGFEER +GKGLI
Sbjct: 1 YLCFGRMTAFSDAQLKEIALGLEASGQNFIWVVKKGLNEKLEWLPEGFEERILGQGKGLI 180
Query: 348 IRGWSPQVMILEHEAIGAFVTHCGWNSVLEGV 379
IRGW+PQVMIL+HE++G FVTHCGWNSVLEGV
Sbjct: 181 IRGWAPQVMILDHESVGGFVTHCGWNSVLEGV 276
Score = 130 bits (327), Expect(2) = 1e-67
Identities = 65/110 (59%), Positives = 84/110 (76%), Gaps = 1/110 (0%)
Frame = +2
Query: 371 GWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKWVMKVG-DNVEWDAV 429
G G VAGVPM+TWP+ +EQFYN K +T+++K GV VGV+ W+ +G D V+ + V
Sbjct: 251 GGTQYWRGCVAGVPMVTWPMYSEQFYNAKFLTDIVKIGVSVGVQTWIGMMGRDPVKKEPV 430
Query: 430 EKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALIEELRS 479
EKAV+R+M GEEA EMRN+AK LA MAK+AVEE GSSY+ N+LIE+LRS
Sbjct: 431 EKAVRRIMVGEEAEEMRNRAKELARMAKRAVEEGGSSYNDFNSLIEDLRS 580
>TC220510 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltransferase,
partial (62%)
Length = 925
Score = 239 bits (610), Expect = 2e-63
Identities = 126/317 (39%), Positives = 186/317 (57%), Gaps = 1/317 (0%)
Frame = +3
Query: 7 PLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNIDI 66
PL + F+ GH IP DMA LF+++G VTI+TTP N + K+L + + +
Sbjct: 6 PLKLYFIHFLAEGHMIPLCDMATLFSTRGHHVTIITTPSNAQILRKSLPSHPL----LRL 173
Query: 67 QTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPHCVVADMFF 126
T++FP E GLP+G EN+ + + + +A +LQ P E+L+ QQ P C+VAD F
Sbjct: 174 HTVQFPSHEVGLPDGIENIYADSDLDSLRKVCSATAMLQPPIEDLVEQQPPDCIVADYLF 353
Query: 127 PWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPGNIKMTR 186
PW D A K IPR+ F+G S F++CA + SSD+ + +
Sbjct: 354 PWVDDLAKKLHIPRLAFNGLSLFTICAIHSSSE-------SSDSTIIQ------------ 476
Query: 187 LQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELE-NVYADYYREVLGIKEW 245
LP+ +T N + K E + ++E++SYG+IVNSF EL+ Y YY + G K W
Sbjct: 477 -SLPHPITLNATPPKELTKFLETVLETELKSYGLIVNSFTELDGEEYTRYYEKTTGHKAW 653
Query: 246 HIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNSQLK 305
H+GP S+ R +E+ + RG+++ + HEC+ WLD+K NSVVY+CFGS+ +F + QL
Sbjct: 654 HLGPASLIGRTAQEK--AERGQKSVVSMHECVAWLDSKRENSVVYICFGSLCYFQDKQLY 827
Query: 306 EIAMGLEASGHNFIWVV 322
EIA G++ASGH+FIWVV
Sbjct: 828 EIACGIQASGHDFIWVV 878
>TC233292 similar to UP|Q8S9A0 (Q8S9A0) Glucosyltransferase-9, partial (46%)
Length = 993
Score = 234 bits (596), Expect = 9e-62
Identities = 120/228 (52%), Positives = 167/228 (72%), Gaps = 11/228 (4%)
Frame = +1
Query: 265 RGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVR- 323
RG +ASI++H CLKWLD + SVVY+CFGS+ + + SQL E+A+ LE + F+WV+R
Sbjct: 214 RGDQASINEHHCLKWLDLQKSKSVVYVCFGSLCNLIPSQLVELALALEDTKRPFVWVIRE 393
Query: 324 -TQTEDGDEWLPE-GFEERTEGKGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVA 381
++ ++ ++W+ E GFEERT+G+GLIIRGW+PQV+IL H AIG F+THCGWNS LEG+ A
Sbjct: 394 GSKYQELEKWISEEGFEERTKGRGLIIRGWAPQVLILSHHAIGGFLTHCGWNSTLEGIGA 573
Query: 382 GVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKWVMKVGDN------VEWDAVEKAVKR 435
G+PMITWP+ A+QF NEKLVT+VLK GV VGV+ MK G+ V+ + + +A+
Sbjct: 574 GLPMITWPLFADQFLNEKLVTKVLKIGVSVGVEV-PMKFGEEEKTGVLVKKEDINRAICM 750
Query: 436 VM--EGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALIEELRSLS 481
VM +GEE+ E R +A L+EMAK+AVE GSS+ L+ LI+++ S
Sbjct: 751 VMDDDGEESKERRERATKLSEMAKRAVENGGSSHLDLSLLIQDIMQQS 894
>TC229865 weakly similar to UP|P93789 (P93789) UDP-glucose
glucosyltransferase , partial (15%)
Length = 647
Score = 211 bits (538), Expect = 5e-55
Identities = 101/191 (52%), Positives = 136/191 (70%), Gaps = 1/191 (0%)
Frame = +1
Query: 1 MDSQSNPLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIH 60
M +++ LH+L FPF +GH IP+ID+A++FAS+G++ T+VTTPLN P IS+ + ++
Sbjct: 85 MGNENRELHVLFFPFPANGHIIPSIDLARVFASRGIKTTVVTTPLNVPLISRTIGKA--- 255
Query: 61 FNNIDIQTIKFPC-VEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLLQQKPHC 119
NI I+TIKFP E GLPEGCEN DS S + F A LL+ P E L+ Q+ P C
Sbjct: 256 --NIKIKTIKFPSHEETGLPEGCENSDSALSSDLIMTFLKATVLLRDPLENLMQQEHPDC 429
Query: 120 VVADMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLP 179
V+ADMF+PWATDSAAKFGIPR+VFHG FF C S C++ Y+P NVSS ++ F + +LP
Sbjct: 430 VIADMFYPWATDSAAKFGIPRVVFHGMGFFPTCVSACVRTYKPQDNVSSWSEPFAVPELP 609
Query: 180 GNIKMTRLQLP 190
G I +T++QLP
Sbjct: 610 GEITITKMQLP 642
>TC218238 weakly similar to UP|Q8H0F2 (Q8H0F2) Anthocyanin
3'-glucosyltransferase, partial (26%)
Length = 936
Score = 206 bits (525), Expect = 1e-53
Identities = 110/209 (52%), Positives = 150/209 (71%), Gaps = 9/209 (4%)
Frame = +3
Query: 281 DTKNINSVVYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTE----DGDEWLPEG 336
D+K NSV+Y+ FGSM F QL EIA LE S H+FIWVVR + E +G+++L E
Sbjct: 3 DSKTENSVLYVSFGSMNKFPTPQLVEIAHALEDSDHDFIWVVRKKGESEDGEGNDFLQE- 179
Query: 337 FEERTEG--KGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQ 394
F++R + KG +I GW+PQ++ILEH AIGA VTHCGWN+++E V AG+PM TWP+ AEQ
Sbjct: 180 FDKRVKASNKGYLIWGWAPQLLILEHHAIGAVVTHCGWNTIIESVNAGLPMATWPLFAEQ 359
Query: 395 FYNEKLVTEVLKTGVPVGVKKW--VMKVGDN-VEWDAVEKAVKRVMEGEEAYEMRNKAKM 451
FYNEKL+ EVL+ GVPVG K+W + GD V+ + + A+ +M GEE+ EMR +AK
Sbjct: 360 FYNEKLLAEVLRIGVPVGAKEWRNWNEFGDEVVKREEIGNAIGVLMGGEESIEMRRRAKA 539
Query: 452 LAEMAKKAVEEDGSSYSQLNALIEELRSL 480
L++ AKKA++ GSS++ L LI+EL+SL
Sbjct: 540 LSDAAKKAIQVGGSSHNNLKELIQELKSL 626
>TC223323 similar to UP|Q8S3B8 (Q8S3B8) Zeatin O-glucosyltransferase, partial
(89%)
Length = 1386
Score = 193 bits (490), Expect = 2e-49
Identities = 145/483 (30%), Positives = 228/483 (47%), Gaps = 16/483 (3%)
Frame = +1
Query: 11 LVFPFMGHGHTIPTIDMAKLFASKGVRVTIV--TTPLNKPPISKALEQSKIHFNNIDIQT 68
++ PF GH + +++L S + V V T + + + S IHF+ ++ +
Sbjct: 49 VLIPFPAQGHLNQLLHLSRLILSHNIPVHYVGTVTHIRQVTLRDHNSISNIHFHAFEVPS 228
Query: 69 IKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELL----LQQKPHCVVADM 124
P P N ++ +P+F A+ L ++P +LL Q K V+ D
Sbjct: 229 FVSP------PPNPNNEETDFPAHLLPSFEASSHL-REPVRKLLHSLSSQAKRVIVIHDS 387
Query: 125 FFPWATDSAAKF-GIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPGNIK 183
A + FH T F K +P D + ++P
Sbjct: 388 VMASVAQDATNMPNVENYTFHSTCTFGTAVFYWDKMGRPL------VDGMLVPEIPSMEG 549
Query: 184 MTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVLGIK 243
N + I+Q + F ++ D G I N+ +E Y ++ G K
Sbjct: 550 CFTTDFMNFM-----IAQ---RDFRKVND------GNIYNTSRAIEGAYIEWMERFTGGK 687
Query: 244 E-WHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNS 302
+ W +GPF+ P K+ S ++H CL+WLD ++ NSV+Y+ FG+ T F
Sbjct: 688 KLWALGPFN----------PLAFEKKDSKERHFCLEWLDKQDPNSVLYVSFGTTTTFKEE 837
Query: 303 QLKEIAMGLEASGHNFIWVVRTQTE----DGDE--W--LPEGFEERTEGKGLIIRGWSPQ 354
Q+K+IA GLE S FIWV+R + DG E W FEER EG GL++R W+PQ
Sbjct: 838 QIKKIATGLEQSKQKFIWVLRDADKGDIFDGSEAKWNEFSNEFEERVEGMGLVVRDWAPQ 1017
Query: 355 VMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVK 414
+ IL H + G F++HCGWNS LE + GVP+ WP+ ++Q N L+TEVLK G+ VK
Sbjct: 1018LEILSHTSTGGFMSHCGWNSCLESISMGVPIAAWPMHSDQPRNSVLITEVLKIGLV--VK 1191
Query: 415 KWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALI 474
W + V VE AV+R+ME +E +MR +A L + ++++E G S ++++ I
Sbjct: 1192NWAQR-NALVSASNVENAVRRLMETKEGDDMRERAVRLKNVIHRSMDEGGVSRMEIDSFI 1368
Query: 475 EEL 477
+
Sbjct: 1369AHI 1377
>BI970958
Length = 789
Score = 193 bits (490), Expect = 2e-49
Identities = 105/229 (45%), Positives = 142/229 (61%), Gaps = 13/229 (5%)
Frame = -2
Query: 265 RGK--EASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVV 322
RGK E +D ECLKWL+TK NSVVY+C GS+ Q EIA G+EASGH F+WV+
Sbjct: 779 RGKXXEDXVDD-ECLKWLNTKESNSVVYICXGSLARLXKEQNXEIARGIEASGHKFLWVL 603
Query: 323 RTQTEDGDE-----WLPEGFEERTEGK--GLIIRGWSPQVMILEHEAIGAFVTHCGWNSV 375
T+D D LP GFEER K G+++RGW PQ +IL+H+AIG F+THCG NSV
Sbjct: 602 PKNTKDDDVKEEELLLPHGFEERMREKKRGMVVRGWVPQGLILKHDAIGGFLTHCGANSV 423
Query: 376 LEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVKKWVMKVGDN----VEWDAVEK 431
+E + GVP+IT P + F EK TEVL GV +GV +W M D V W+ +E
Sbjct: 422 VEAICEGVPLITMPRFGDHFLCEKQATEVLGLGVELGVSEWSMSPYDARKEVVGWERIEN 243
Query: 432 AVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALIEELRSL 480
AV++VM+ +E + + K + E A + V+E G+SY + L++ LR +
Sbjct: 242 AVRKVMK-DEGGLLNKRVKEMKEKAHEVVQEGGNSYDNVTTLVQSLRRM 99
>TC213945 similar to UP|Q6QDB6 (Q6QDB6) UDP-glucose glucosyltransferase,
partial (23%)
Length = 475
Score = 192 bits (489), Expect = 2e-49
Identities = 95/162 (58%), Positives = 121/162 (74%), Gaps = 1/162 (0%)
Frame = +2
Query: 134 AKFGIPRIVFHGTSFFSLCASQCMKKYQPY-KNVSSDTDLFEITDLPGNIKMTRLQLPNT 192
AKFGIPR+VFHGTSFFSLC + CM Y+P+ K SSD+D F I + PG I++ + ++P
Sbjct: 2 AKFGIPRLVFHGTSFFSLCVTTCMPFYEPHDKYASSDSDSFLIPNFPGEIRIEKTKIPPY 181
Query: 193 LTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVLGIKEWHIGPFSI 252
+ AKL EE K+SE+RSYGV+VNSFYELE VYAD++R VLG K WHIGP S+
Sbjct: 182 SKSKE--KAGLAKLLEEAKESELRSYGVVVNSFYELEKVYADHFRNVLGRKAWHIGPLSL 355
Query: 253 HNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFG 294
N++ EE+ + RGKEASID+HECLKWL+TK NSV+Y+C G
Sbjct: 356 CNKDAEEK--ARRGKEASIDEHECLKWLNTKKPNSVIYICXG 475
>TC227846 weakly similar to UP|Q947K4 (Q947K4) Thiohydroximate
S-glucosyltransferase, partial (49%)
Length = 1605
Score = 191 bits (485), Expect = 6e-49
Identities = 149/480 (31%), Positives = 235/480 (48%), Gaps = 13/480 (2%)
Frame = +2
Query: 11 LVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNIDIQTIK 70
LV P+ GH P + AK ASKGV+ T+ TT + I+ NI ++ I
Sbjct: 2 LVLPYPAQGHINPLVQFAKRLASKGVKATVATTHYTA---------NSINAPNITVEAIS 154
Query: 71 FPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLL-QQPFEELLL--QQKPH---CVVADM 124
+AG + NV F A+ R + EL+ QQ P C+V D
Sbjct: 155 DGFDQAGFAQTNNNVQ---------LFLASFRTNGSRTLSELIRKHQQTPSPSTCIVYDS 307
Query: 125 FFPWATDSAAKFGIPRIVFHGTSFFSLCASQC---MKKYQPYKNVSSDTDLFEITDLPGN 181
FFPW D A + GI +G +FF+ A+ C + + + + + + +PG
Sbjct: 308 FFPWVLDVAKQHGI-----YGAAFFTNSAAVCNIFCRLHHGFIQLPVKMEHLPLR-VPGL 469
Query: 182 IKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVLG 241
+ LP+ + + A + + + + VN+F LE+ E+
Sbjct: 470 PPLDSRALPSFVRFPESYPAYMAMKLSQFSNLNNADW-MFVNTFEALESEVLKGLTELFP 646
Query: 242 IKEWHIGPFSIHNRNKEEEIPSYRGKEASIDK---HECLKWLDTKNINSVVYMCFGSMTH 298
K IGP + + + I +G AS+ K EC WL++K SVVY+ FGSM
Sbjct: 647 AKM--IGPM-VPSGYLDGRIKGDKGYGASLWKPLTEECSNWLESKPPQSVVYISFGSMVS 817
Query: 299 FLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDEWLPEGFEERTEGKGLIIRGWSPQVMIL 358
Q++E+A GL+ SG +F+WV+R ++E G LP G+ E + KGLI+ W Q+ +L
Sbjct: 818 LTEEQMEEVAWGLKESGVSFLWVLR-ESEHGK--LPLGYRESVKDKGLIVT-WCNQLELL 985
Query: 359 EHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGV-PVGVKKWV 417
H+A G FVTHCGWNS LE + GVP++ P A+Q + K + E+ + GV P +K +
Sbjct: 986 AHQATGCFVTHCGWNSTLESLSLGVPVVCLPQWADQLPDAKFLDEIWEVGVWPKEDEKGI 1165
Query: 418 MKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALIEEL 477
++ + V+ ++K VMEG+ + E+R A ++A++AV E GSS +N ++ L
Sbjct: 1166VRKQEFVQ------SLKDVMEGQRSQEIRRNANKWKKLAREAVGEGGSSDKHINQFVDHL 1327
>NP600052 zeatin O-glucosyltransferase [Glycine max]
Length = 1395
Score = 189 bits (479), Expect = 3e-48
Identities = 146/483 (30%), Positives = 216/483 (44%), Gaps = 15/483 (3%)
Frame = +1
Query: 10 ILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNIDIQTI 69
+++ PF GH + +A+ S + V V T + + S I I
Sbjct: 40 VVLIPFPAQGHLNQLLHLARHIFSHNIPVHYVGTATHIRQATLRDHNSNISNIIIHFHAF 219
Query: 70 KFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELLL----QQKPHCVVADMF 125
+ P + P PS +P+F A+ L ++P LL Q K V+ D
Sbjct: 220 EVPPFVSPPPNPNNEETDFPS-HLLPSFKASSHL-REPVRNLLQSLSSQAKRVIVIHDSL 393
Query: 126 FPWATDSAAKF-GIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPGNIKM 184
A + FH T CA Y F+ T++P
Sbjct: 394 MASVAQDATNMPNVENYTFHST-----CAFTTFLYYWEVMGRPPVEGFFQATEIPSMGGC 558
Query: 185 TRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVLGIKE 244
Q + +TE F + D G I N+ +E Y ++ + G K+
Sbjct: 559 FPPQFIHFITEEYE--------FHQFND------GNIYNTSRAIEGPYIEFLERIGGSKK 696
Query: 245 --WHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNS 302
W +GPF+ P K+ +H C++WLD + NSV+Y+ FG+ T F +
Sbjct: 697 RLWALGPFN----------PLTIEKKDPKTRHICIEWLDKQEANSVMYVSFGTTTSFTVA 846
Query: 303 QLKEIAMGLEASGHNFIWVVRTQTE----DGDEW----LPEGFEERTEGKGLIIRGWSPQ 354
Q ++IA+GLE S FIWV+R + DG E LP GFEER EG GL+IR W+PQ
Sbjct: 847 QFEQIAIGLEQSKQKFIWVLRDADKGNIFDGSEAERYELPNGFEERVEGIGLLIRDWAPQ 1026
Query: 355 VMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGVPVGVK 414
+ IL H + G F++HCGWNS LE + GVP+ WP+ ++Q N L+TEVLK G VK
Sbjct: 1027LEILSHTSTGGFMSHCGWNSCLESITMGVPIAAWPMHSDQPRNSVLITEVLKVGFV--VK 1200
Query: 415 KWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQLNALI 474
W + V VE AV+R+ME +E EMR++A L ++ G S ++ + I
Sbjct: 1201DWAQR-NALVSASVVENAVRRLMETKEGDEMRDRAVRLKNCIHRSKYGGGVSRMEMGSFI 1377
Query: 475 EEL 477
+
Sbjct: 1378AHI 1386
>TC218577 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (Fragment),
partial (58%)
Length = 1530
Score = 185 bits (470), Expect = 3e-47
Identities = 149/493 (30%), Positives = 229/493 (46%), Gaps = 19/493 (3%)
Frame = +1
Query: 9 HILVFPFMGHGHTIPTIDMAK--LFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNIDI 66
H+ V P G H +P ++ +K L +T + P S SK + +
Sbjct: 4 HVAVVPSPGFTHLVPILEFSKRLLHLHPEFHITCFIPSVGSSPTS-----SKAY-----V 153
Query: 67 QTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFA---AIRLLQQPFEELLLQQKPHCVVAD 123
QT+ LP + S PSV + + ++ +++ + L + K +V D
Sbjct: 154 QTLPPTITSIFLPPITLDHVSDPSVLALQIELSVNLSLPYIREELKSLCSRAKVVALVVD 333
Query: 124 MFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPGNIK 183
+F A + A + + ++ S L K + S +L + D+PG +
Sbjct: 334 VFANGALNFAKELNLLSYIYLPQSAMLLSLYFYSTKLDEILS-SESRELQKPIDIPGCVP 510
Query: 184 MTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVLGIK 243
+ LP L +D + E K V GV +N+F ELE+ E + K
Sbjct: 511 IHNKDLP--LPFHDLSGLGYKGFLERSKRFHVPD-GVFMNTFLELESGAIRALEEHVKGK 681
Query: 244 E--WHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLN 301
+ +GP + E I G E + ECL WLD + NSV+Y+ FGS
Sbjct: 682 PKLYPVGPII-----QMESI----GHENGV---ECLTWLDKQEPNSVLYVSFGSGGTLSQ 825
Query: 302 SQLKEIAMGLEASGHNFIWVVRT------------QTEDGDEWLPEGFEERTEGKGLIIR 349
Q E+A GLE SG F+WVVR +T+D E+LP GF ERT+ +GL++
Sbjct: 826 EQFNELAFGLELSGKKFLWVVRAPSGVVSAGYLCAETKDPLEFLPHGFLERTKKQGLVVP 1005
Query: 350 GWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEVLKTGV 409
W+PQ+ +L H A G F++HCGWNSVLE VV GVP+ITWP+ AEQ N ++ + LK +
Sbjct: 1006SWAPQIQVLGHSATGGFLSHCGWNSVLESVVQGVPVITWPLFAEQSLNAAMIADDLKVAL 1185
Query: 410 PVGVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDGSSYSQ 469
V + VE + + K V+ +M +E+ E+R + +L A A++EDGSS
Sbjct: 1186RPKVNE-----SGLVEREEIAKVVRGLMGDKESLEIRKRMGLLKIAAANAIKEDGSSTKT 1350
Query: 470 LNALIEELRSLSH 482
L+ + LR H
Sbjct: 1351LSEMATSLRGF*H 1389
>CO986104
Length = 797
Score = 177 bits (450), Expect = 7e-45
Identities = 96/249 (38%), Positives = 143/249 (56%), Gaps = 2/249 (0%)
Frame = +1
Query: 5 SNPLHILVFPFMGHGHTIPTIDMAKLFASKGVRVTIVTTPLNKPPISKALEQSKIHFNNI 64
SN H ++FP M GH IP +D+A+L A +GV VTI TTP N + L ++ I
Sbjct: 55 SNNPHFVLFPLMAQGHIIPMMDIARLLARRGVIVTIFTTPKNASRFNSVLSRAVSSGLQI 234
Query: 65 DIQTIKFPCVEAGLPEGCENVDSIPSVSFVPAFFAAIRLLQQPFEELL--LQQKPHCVVA 122
+ + FP EAGLPEGCEN D + S+ + F AI +LQ+ EEL L KP C+++
Sbjct: 235 RLVQLHFPSKEAGLPEGCENFDMLTSMDMMYKVFHAISMLQKSAEELFEALIPKPSCIIS 414
Query: 123 DMFFPWATDSAAKFGIPRIVFHGTSFFSLCASQCMKKYQPYKNVSSDTDLFEITDLPGNI 182
D PW A K IPRI FHG S F L + ++++S+++ F I +PG I
Sbjct: 415 DFCIPWTAQVAEKHHIPRISFHGFSCFCLHCLLMVHTSNICESITSESEYFTIPGIPGQI 594
Query: 183 KMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYELENVYADYYREVLGI 242
+ T+ Q+P ++ +D + F ++++D+E++SYG+I+N+F EL Y Y++V
Sbjct: 595 QATKEQIPMMISNSDEEMKHFG---DQMRDAEMKSYGLIINTFEELXXAYVTDYKKVRND 765
Query: 243 KEWHIGPFS 251
K W IGP S
Sbjct: 766 KVWCIGPVS 792
>TC203711 similar to UP|Q8S9A4 (Q8S9A4) Glucosyltransferase-5, partial (57%)
Length = 1322
Score = 177 bits (449), Expect = 9e-45
Identities = 116/316 (36%), Positives = 170/316 (53%), Gaps = 11/316 (3%)
Frame = +3
Query: 169 DTDLFEITDLPGNIKMTRLQLPNTLTENDPISQSFAKLFEEIKDSEVRSYGVIVNSFYEL 228
DTD +PG +T PN DP+S + ++F +I ++ + G+IVN+F +
Sbjct: 195 DTDQPLQIQIPGLSTITADDFPNEC--KDPLSYA-CQVFLQIAETMMGGAGIIVNTFEAI 365
Query: 229 ENVYADYYREVLGIKE--WHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNIN 286
E E + + +GP I + G+E DK CL WL+ +
Sbjct: 366 EEEAIRALSEDATVPPPLFCVGPV----------ISAPYGEE---DKG-CLSWLNLQPSQ 503
Query: 287 SVVYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQ---TEDG------DEWLPEGF 337
SVV +CFGSM F +QLKEIA+GLE S F+WVVRT+ +D DE LPEGF
Sbjct: 504 SVVLLCFGSMGRFSRAQLKEIAIGLEKSEQRFLWVVRTELGGADDSAEELSLDELLPEGF 683
Query: 338 EERTEGKGLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYN 397
ERT+ KG+++R W+PQ IL H+++G FVTHCGWNSVLE V GVPM+ WP+ AEQ N
Sbjct: 684 LERTKEKGMVVRDWAPQAAILSHDSVGGFVTHCGWNSVLEAVCEGVPMVAWPLYAEQKMN 863
Query: 398 EKLVTEVLKTGVPVGVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAK 457
++ + +K + V K V + V+ +ME ++ E+R + + A
Sbjct: 864 RMVMVKEMKVALAVKENK-----DGFVSSTELGDRVRELMESDKGKEIRQRIFKMKMSAA 1028
Query: 458 KAVEEDGSSYSQLNAL 473
+A+ E G+S + L+ L
Sbjct: 1029EAMAEGGTSRASLDKL 1076
>TC210204 similar to UP|Q9SY84 (Q9SY84) F14N23.30, partial (21%)
Length = 782
Score = 174 bits (442), Expect = 6e-44
Identities = 92/203 (45%), Positives = 131/203 (64%), Gaps = 1/203 (0%)
Frame = +3
Query: 286 NSVVYMCFGSMTHFLNSQLKEIAMGLEASGHNFIWVVRTQTEDGDEW-LPEGFEERTEGK 344
+SV+Y FGS QL+EIA GLE S +F+WV+R + EW LP+G+EER + +
Sbjct: 42 SSVLYAAFGSQAEISREQLEEIAKGLEESKVSFLWVIRKE-----EWGLPDGYEERVKDR 206
Query: 345 GLIIRGWSPQVMILEHEAIGAFVTHCGWNSVLEGVVAGVPMITWPVAAEQFYNEKLVTEV 404
G++IR W Q IL HE++ F++HCGWNSV+E V AGVP++ WP+ AEQF N ++V E
Sbjct: 207 GIVIREWVDQREILMHESVEGFLSHCGWNSVMESVTAGVPIVGWPIMAEQFLNARMVEEE 386
Query: 405 LKTGVPVGVKKWVMKVGDNVEWDAVEKAVKRVMEGEEAYEMRNKAKMLAEMAKKAVEEDG 464
+K G + V+ V V+ + ++K VK VMEG + ++R K + LAEMAK A +E G
Sbjct: 387 VKVG--LRVETCDGSVRGFVKREGLKKTVKEVMEGVKGKKLREKVRELAEMAKLATQEGG 560
Query: 465 SSYSQLNALIEELRSLSHHQHIS 487
SS S LN+L+ + + SH IS
Sbjct: 561 SSCSTLNSLLHQTCAASHKNQIS 629
>BU926438 similar to PIR|T03747|T03 glucosyltransferase IS5a (EC 2.4.1.-)
salicylate-induced - common tobacco, partial (21%)
Length = 446
Score = 172 bits (437), Expect = 2e-43
Identities = 78/148 (52%), Positives = 105/148 (70%), Gaps = 7/148 (4%)
Frame = +1
Query: 245 WHIGPFSIHNRNKEEEIPSYRGKEASIDKHECLKWLDTKNINSVVYMCFGSMTHFLNSQL 304
WH+GP S+ + +E + RG ++++ +C+ WLD+K +NSV+Y+CFGS+ HF + QL
Sbjct: 4 WHLGPASLISCRTAQE-KAERGMKSAVSMQDCVSWLDSKRVNSVLYICFGSLCHFPDEQL 180
Query: 305 KEIAMGLEASGHNFIWVV-------RTQTEDGDEWLPEGFEERTEGKGLIIRGWSPQVMI 357
EIA G+EASGH FIWVV E+ ++WLP GFEER KG+IIRGW+PQV+I
Sbjct: 181 YEIACGMEASGHEFIWVVPEKKGKEHESEEEKEKWLPRGFEERNAEKGMIIRGWAPQVII 360
Query: 358 LEHEAIGAFVTHCGWNSVLEGVVAGVPM 385
L H A+GAF+THCGWNS +E V GVPM
Sbjct: 361 LGHPAVGAFITHCGWNSTVEAVSEGVPM 444
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,677,203
Number of Sequences: 63676
Number of extensions: 324574
Number of successful extensions: 1833
Number of sequences better than 10.0: 199
Number of HSP's better than 10.0 without gapping: 1664
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1673
length of query: 489
length of database: 12,639,632
effective HSP length: 101
effective length of query: 388
effective length of database: 6,208,356
effective search space: 2408842128
effective search space used: 2408842128
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC149574.3


