

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149574.1 - phase: 0 /pseudo
(619 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
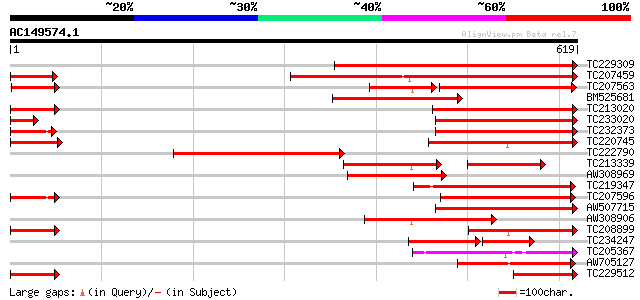
Score E
Sequences producing significant alignments: (bits) Value
TC229309 homologue to UP|Q9LJX0 (Q9LJX0) P-glycoprotein; multi-d... 315 3e-86
TC207459 similar to UP|Q94IH6 (Q94IH6) CjMDR1, partial (30%) 306 2e-83
TC207563 similar to UP|Q9ZRG2 (Q9ZRG2) P-glycoprotein, partial (... 212 2e-67
BM525681 similar to GP|6671365|gb P-glycoprotein {Gossypium hirs... 236 2e-62
TC213020 weakly similar to UP|Q8GU71 (Q8GU71) MDR-like ABC trans... 224 1e-58
TC233020 similar to UP|Q9LHD1 (Q9LHD1) Multidrug resistance p-gl... 205 4e-53
TC232373 weakly similar to UP|AB10_MOUSE (Q9JI39) ATP-binding ca... 191 6e-49
TC220745 similar to UP|Q9LHD1 (Q9LHD1) Multidrug resistance p-gl... 190 2e-48
TC222790 181 7e-46
TC213339 similar to UP|Q9ZRG2 (Q9ZRG2) P-glycoprotein, partial (... 121 2e-44
AW308969 similar to GP|6671365|gb P-glycoprotein {Gossypium hirs... 174 1e-43
TC219347 similar to UP|Q9LF78 (Q9LF78) Mitochondrial half-ABC tr... 172 3e-43
TC207596 similar to UP|Q7GBY1 (Q7GBY1) Tonoplast ABC transporter... 164 8e-41
AW507715 weakly similar to GP|9294227|db P-glycoprotein; multi-d... 159 3e-39
AW308906 154 9e-38
TC208899 similar to UP|Q9LHD1 (Q9LHD1) Multidrug resistance p-gl... 153 2e-37
TC234247 similar to GB|CAD59589.1|27368863|OSA535067 MDR-like AB... 90 1e-30
TC205367 similar to UP|O24525 (O24525) Glutathione-conjugate tra... 113 2e-25
AW705127 weakly similar to GP|27359084|gb ABC-type bacteriocin/l... 108 7e-24
TC229512 similar to UP|Q9SDM5 (Q9SDM5) P-glycoprotein, partial (... 106 3e-23
>TC229309 homologue to UP|Q9LJX0 (Q9LJX0) P-glycoprotein; multi-drug
resistance related; ABC transporter-like protein,
partial (22%)
Length = 841
Score = 315 bits (808), Expect = 3e-86
Identities = 156/267 (58%), Positives = 214/267 (79%), Gaps = 2/267 (0%)
Frame = +1
Query: 355 SQFFIFSSYGLALWYGSVLLEKELASFKSIMKSFMVLIVTALAMGETLALAPDLLKGNQM 414
SQ +++S L LWYG+ L+ K +++F ++K F+VL++TA ++ ET++LAP++++G +
Sbjct: 1 SQLALYASEALILWYGAHLVSKGVSTFSKVIKVFVVLVITANSVAETVSLAPEIIRGGEA 180
Query: 415 VSSIFDMIDRKSGIIHDV--GEELMTVEGMIELKRINFIYPSRPNVVIFKDFNLIVPSGK 472
V S+F ++DR + I D + + ++ G IEL+ ++F YPSRP+V++FKD NL + +G+
Sbjct: 181 VGSVFSILDRSTRIDPDDPDADPVESLRGEIELRHVDFAYPSRPDVMVFKDLNLRIRAGQ 360
Query: 473 SLALVGHSGSGKSSIISLILRFYDPTSGKVMIDGKDIKKMNLKSLRKQIGLVQQEPALFA 532
S ALVG SGSGKSS+I+LI RFYDP +GKVM+DGKDI+K+NLKSLR +IGLVQQEPALFA
Sbjct: 361 SQALVGASGSGKSSVIALIERFYDPIAGKVMVDGKDIRKLNLKSLRLKIGLVQQEPALFA 540
Query: 533 TSIYKNILYGKEEASESEVIEAAKLADAHNFISALPEGYSTKAGDRGVLLSGGQKQRVAI 592
SI++NI YGKE A+E+EVIEAA+ A+ H F+S LPEGY T G+RGV LSGGQKQR+AI
Sbjct: 541 ASIFENIAYGKEGATEAEVIEAARAANVHGFVSGLPEGYKTPVGERGVQLSGGQKQRIAI 720
Query: 593 ARAILRNPKILLLDEATSALDVESERV 619
ARA+L++P ILLLDEATSALD ESE V
Sbjct: 721 ARAVLKDPTILLLDEATSALDAESECV 801
>TC207459 similar to UP|Q94IH6 (Q94IH6) CjMDR1, partial (30%)
Length = 1329
Score = 306 bits (783), Expect = 2e-83
Identities = 159/318 (50%), Positives = 231/318 (72%), Gaps = 5/318 (1%)
Frame = +1
Query: 307 AGEAVSNIRTVAAFCAEEKVIDLYADELVEPSKRSFKRGQIAGIFYGISQFFIFSSYGLA 366
+ +AV +IRTVA+FCAE+KV++LY ++ P K ++G I+G +G+S F +F Y +
Sbjct: 4 SNDAVGSIRTVASFCAEDKVMELYKNKCEGPMKTGIRQGLISGSGFGVSFFLLFCVYATS 183
Query: 367 LWYGSVLLEKELASFKSIMKSFMVLIVTALAMGETLALAPDLLKGNQMVSSIFDMIDRKS 426
+ G+ L++ A+F + + F L + A+ + ++ + APD K +SIF +ID+KS
Sbjct: 184 FYAGARLVDAGKATFSDVFRVFFALTMAAIGVSQSSSFAPDSSKAKSATASIFGIIDKKS 363
Query: 427 GIIHDVGEE----LMTVEGMIELKRINFIYPSRPNVVIFKDFNLIVPSGKSLALVGHSGS 482
I D G+E L +V+G IEL+ ++F YPSRP++ IF+D +L + SGK++ALVG SGS
Sbjct: 364 KI--DPGDESGSTLDSVKGEIELRHVSFKYPSRPDIQIFRDLSLTIHSGKTVALVGESGS 537
Query: 483 GKSSIISLILRFYDPTSGKVMIDGKDIKKMNLKSLRKQIGLVQQEPALFATSIYKNILYG 542
GKS++I+L+ RFY+P SG++ +DG +I+++ LK LR+Q+GLV QEP LF +I NI YG
Sbjct: 538 GKSTVIALLQRFYNPDSGQITLDGIEIRELQLKWLRQQMGLVSQEPVLFNETIRANIAYG 717
Query: 543 K-EEASESEVIEAAKLADAHNFISALPEGYSTKAGDRGVLLSGGQKQRVAIARAILRNPK 601
K +A+E+E+I AA++A+AH FIS L +GY T G+RG LSGGQKQRVAIARAI+++PK
Sbjct: 718 KGGDATEAEIIAAAEMANAHKFISGLQQGYDTIVGERGTQLSGGQKQRVAIARAIIKSPK 897
Query: 602 ILLLDEATSALDVESERV 619
ILLLDEATSALD ESERV
Sbjct: 898 ILLLDEATSALDAESERV 951
Score = 70.9 bits (172), Expect = 2e-12
Identities = 34/52 (65%), Positives = 42/52 (80%)
Frame = +1
Query: 1 MIGRTTVIVAHRLSTIKNADMIAVVEGGSVVETGNHEELISNPNSLYSSLVQ 52
M+ RTTV+VAHRLSTIKNAD+IAVV+ G +VE G HE+LI+ Y+SLVQ
Sbjct: 976 MVNRTTVVVAHRLSTIKNADVIAVVKNGVIVEKGKHEKLINVSGGFYASLVQ 1131
>TC207563 similar to UP|Q9ZRG2 (Q9ZRG2) P-glycoprotein, partial (24%)
Length = 1164
Score = 212 bits (540), Expect(2) = 2e-67
Identities = 106/149 (71%), Positives = 127/149 (85%)
Frame = +1
Query: 470 SGKSLALVGHSGSGKSSIISLILRFYDPTSGKVMIDGKDIKKMNLKSLRKQIGLVQQEPA 529
+GK+LALVG SG GKSSII+LI RFYDPTSG+VMIDGKDI+K NLKSLR+ I +V QEP
Sbjct: 247 AGKTLALVGPSGCGKSSIIALIQRFYDPTSGRVMIDGKDIRKYNLKSLRRHISVVPQEPC 426
Query: 530 LFATSIYKNILYGKEEASESEVIEAAKLADAHNFISALPEGYSTKAGDRGVLLSGGQKQR 589
LFAT+IY+NI YG E A+E+E+IEAA LA+AH FIS LP+GY T G+RGV LSGGQKQR
Sbjct: 427 LFATTIYENIAYGHESATEAEIIEAATLANAHKFISGLPDGYKTFVGERGVQLSGGQKQR 606
Query: 590 VAIARAILRNPKILLLDEATSALDVESER 618
+A+ARA +R +++LLDEATSALD ESER
Sbjct: 607 IAVARAFVRKAELMLLDEATSALDAESER 693
Score = 63.5 bits (153), Expect = 3e-10
Identities = 28/53 (52%), Positives = 43/53 (80%), Gaps = 1/53 (1%)
Frame = +1
Query: 3 GRTTVIVAHRLSTIKNADMIAVVEGGSVVETGNHEELISN-PNSLYSSLVQGQ 54
G+TT+IVAHRLSTI+NA++IAV++ G V E G+H +L+ N P+ +Y+ ++Q Q
Sbjct: 727 GKTTIIVAHRLSTIRNANLIAVIDDGKVAEQGSHSQLLKNHPDGIYARMIQLQ 885
Score = 62.8 bits (151), Expect(2) = 2e-67
Identities = 30/77 (38%), Positives = 50/77 (63%), Gaps = 3/77 (3%)
Frame = +3
Query: 393 VTALAMGETLALAPDLLKGNQMVSSIFDMIDRKSGIIHDVGEELMT---VEGMIELKRIN 449
V+A ETL LAPD +KG Q + S+F+++DR++ I D + + G +ELK ++
Sbjct: 6 VSANGAAETLTLAPDFIKGGQAMRSVFELLDRRTEIEPDDQDATPVPDRLRGEVELKHVD 185
Query: 450 FIYPSRPNVVIFKDFNL 466
F YP+RP++ +F+D +L
Sbjct: 186 FSYPTRPDMPVFRDLSL 236
>BM525681 similar to GP|6671365|gb P-glycoprotein {Gossypium hirsutum},
partial (11%)
Length = 428
Score = 236 bits (603), Expect = 2e-62
Identities = 120/142 (84%), Positives = 134/142 (93%)
Frame = +2
Query: 353 GISQFFIFSSYGLALWYGSVLLEKELASFKSIMKSFMVLIVTALAMGETLALAPDLLKGN 412
GISQFFIFSSYGLALWYGSVL+EKELASFKSIMK+F VLIVTALAMGETLALAPDLLKGN
Sbjct: 2 GISQFFIFSSYGLALWYGSVLMEKELASFKSIMKAFFVLIVTALAMGETLALAPDLLKGN 181
Query: 413 QMVSSIFDMIDRKSGIIHDVGEELMTVEGMIELKRINFIYPSRPNVVIFKDFNLIVPSGK 472
QMV+S+F+++DRKSGI +VGEEL TV+G IELKRINF YPSRP+V+IFKDFNL VP+GK
Sbjct: 182 QMVASVFEVMDRKSGISCEVGEELKTVDGTIELKRINFSYPSRPDVIIFKDFNLRVPAGK 361
Query: 473 SLALVGHSGSGKSSIISLILRF 494
S+ALVG SGSGKSS+ISLILRF
Sbjct: 362 SVALVGQSGSGKSSVISLILRF 427
>TC213020 weakly similar to UP|Q8GU71 (Q8GU71) MDR-like ABC transporter,
partial (18%)
Length = 864
Score = 224 bits (570), Expect = 1e-58
Identities = 112/158 (70%), Positives = 138/158 (86%)
Frame = +1
Query: 462 KDFNLIVPSGKSLALVGHSGSGKSSIISLILRFYDPTSGKVMIDGKDIKKMNLKSLRKQI 521
++ NL VP+GKSLA+VG SGSGKS++ISL++RFYDP SG V++D DIK +NL+SLR +I
Sbjct: 1 ENLNLRVPAGKSLAVVGQSGSGKSTVISLVMRFYDPDSGLVLVDECDIKNLNLRSLRLRI 180
Query: 522 GLVQQEPALFATSIYKNILYGKEEASESEVIEAAKLADAHNFISALPEGYSTKAGDRGVL 581
GLVQQEPALF+T++Y+NI YGKEEASE EV++AAK A+AH FIS +PEGY T+ G+RGV
Sbjct: 181 GLVQQEPALFSTTVYENIKYGKEEASEIEVMKAAKAANAHEFISRMPEGYKTEVGERGVQ 360
Query: 582 LSGGQKQRVAIARAILRNPKILLLDEATSALDVESERV 619
LSGGQKQRVAIARAIL++P ILLLDEATSALD SER+
Sbjct: 361 LSGGQKQRVAIARAILKDPSILLLDEATSALDTVSERL 474
Score = 61.2 bits (147), Expect = 1e-09
Identities = 28/54 (51%), Positives = 40/54 (73%)
Frame = +1
Query: 1 MIGRTTVIVAHRLSTIKNADMIAVVEGGSVVETGNHEELISNPNSLYSSLVQGQ 54
M GRTT++VAHRLST+++A+ IAV++ G V E G+HE L++ S+Y LV Q
Sbjct: 499 MEGRTTILVAHRLSTVRDANSIAVLQNGRVAEMGSHERLMAKSGSIYKQLVSLQ 660
>TC233020 similar to UP|Q9LHD1 (Q9LHD1) Multidrug resistance p-glycoprotein,
partial (15%)
Length = 605
Score = 205 bits (522), Expect = 4e-53
Identities = 102/154 (66%), Positives = 128/154 (82%)
Frame = +1
Query: 466 LIVPSGKSLALVGHSGSGKSSIISLILRFYDPTSGKVMIDGKDIKKMNLKSLRKQIGLVQ 525
L VP+GK +ALVG SGSGKS++I+L+ RFYDP G+V +DG I+K+ LK LR +GLV
Sbjct: 25 LRVPAGKRVALVGESGSGKSTVIALLQRFYDPCGGEVRVDGVGIQKLQLKWLRSCMGLVS 204
Query: 526 QEPALFATSIYKNILYGKEEASESEVIEAAKLADAHNFISALPEGYSTKAGDRGVLLSGG 585
QEPALFATSI NIL+GKE+A++ +V+EAAK A AHNFIS LP GY T+ G+RG+ +SGG
Sbjct: 205 QEPALFATSIKDNILFGKEDATQDQVVEAAKAAHAHNFISLLPHGYHTQVGERGIQMSGG 384
Query: 586 QKQRVAIARAILRNPKILLLDEATSALDVESERV 619
QKQR+AIARAI++ P+ILLLDEATSALD ESER+
Sbjct: 385 QKQRIAIARAIIKKPRILLLDEATSALDSESERL 486
Score = 47.4 bits (111), Expect = 2e-05
Identities = 20/30 (66%), Positives = 27/30 (89%)
Frame = +1
Query: 2 IGRTTVIVAHRLSTIKNADMIAVVEGGSVV 31
+G TT+I+AHRLSTI+NAD+IAVV GG ++
Sbjct: 514 VGCTTIIIAHRLSTIQNADLIAVVGGGKII 603
>TC232373 weakly similar to UP|AB10_MOUSE (Q9JI39) ATP-binding cassette,
sub-family B, member 10, mitochondrial precursor
(ATP-binding cassette transporter 10) (ABC transporter
10 protein) (ABC-mitochondrial erythroid protein)
(ABC-me protein), partial (24%)
Length = 742
Score = 191 bits (486), Expect = 6e-49
Identities = 97/155 (62%), Positives = 127/155 (81%), Gaps = 1/155 (0%)
Frame = +1
Query: 466 LIVPSGKSLALVGHSGSGKSSIISLILRFYDPTSGKVMIDGKDIKKMNLKSLRKQIGLVQ 525
L + +G+++AL G SGSGKS++ISL+ RFY+P SG++ +DG +I+K+ LK R+Q+GLV
Sbjct: 1 LTIHAGETVALAGESGSGKSTVISLLQRFYEPDSGQITLDGTEIQKLQLKWFRQQMGLVS 180
Query: 526 QEPALFATSIYKNILYGKE-EASESEVIEAAKLADAHNFISALPEGYSTKAGDRGVLLSG 584
QEP LF +I NI YGK +A+E+E+I A +LA+AH FIS+L +GY T G+RG+ LSG
Sbjct: 181 QEPVLFNDTIRTNIAYGKGGDATEAEIIAATELANAHTFISSLQQGYDTIVGERGIQLSG 360
Query: 585 GQKQRVAIARAILRNPKILLLDEATSALDVESERV 619
GQKQRVAIARAI++NPKILLLDEATSALDVESERV
Sbjct: 361 GQKQRVAIARAIVKNPKILLLDEATSALDVESERV 465
Score = 60.8 bits (146), Expect = 2e-09
Identities = 29/51 (56%), Positives = 39/51 (75%)
Frame = +1
Query: 1 MIGRTTVIVAHRLSTIKNADMIAVVEGGSVVETGNHEELISNPNSLYSSLV 51
M+ RTT++VAHRLSTIK+AD IAVV+ G + E G H+ L+ N +Y+SLV
Sbjct: 490 MVDRTTIVVAHRLSTIKDADSIAVVQNGVIAEQGKHDTLL-NKGGIYASLV 639
>TC220745 similar to UP|Q9LHD1 (Q9LHD1) Multidrug resistance p-glycoprotein,
partial (18%)
Length = 1050
Score = 190 bits (482), Expect = 2e-48
Identities = 98/166 (59%), Positives = 127/166 (76%), Gaps = 4/166 (2%)
Frame = +3
Query: 458 VVIFKDFNLIVPSGKSLALVGHSGSGKSSIISLILRFYDPTSGKVMIDGKDIKKMNLKSL 517
V+IF+ F++ + + +S ALVG SGSG S+II LI RFYDP G V IDG+DIK +L+SL
Sbjct: 3 VMIFQGFSIXIDAXRSTALVGQSGSGXSTIIGLIERFYDPMKGIVTIDGRDIKSYHLRSL 182
Query: 518 RKQIGLVQQEPALFATSIYKNILYG----KEEASESEVIEAAKLADAHNFISALPEGYST 573
RK I LV QEP LF +I +NI YG + E+E+IEAA+ A+AH+FI++L +GY T
Sbjct: 183 RKHIALVSQEPTLFGGTIRENIAYGASNNNNKVDETEIIEAARAANAHDFIASLKDGYDT 362
Query: 574 KAGDRGVLLSGGQKQRVAIARAILRNPKILLLDEATSALDVESERV 619
DRGV LSGGQKQR+AIARAIL+NP++LLLDEATSALD +SE++
Sbjct: 363 SCRDRGVQLSGGQKQRIAIARAILKNPEVLLLDEATSALDSQSEKL 500
Score = 63.5 bits (153), Expect = 3e-10
Identities = 31/58 (53%), Positives = 42/58 (71%), Gaps = 1/58 (1%)
Frame = +3
Query: 1 MIGRTTVIVAHRLSTIKNADMIAVVEGGSVVETGNHEELISN-PNSLYSSLVQGQPSP 57
M+GRT+V+VAHRLSTI+N D+IAV++ G VVE G H L+++ P Y SL+ Q P
Sbjct: 525 MVGRTSVVVAHRLSTIQNCDLIAVLDKGKVVEKGTHSSLLAHGPGGAYYSLISLQRRP 698
>TC222790
Length = 563
Score = 181 bits (460), Expect = 7e-46
Identities = 91/186 (48%), Positives = 127/186 (67%)
Frame = +1
Query: 180 YSIEHLSFGIMGERLTLRVRGIMLSAILKNEIGWFDDTRNTSSMLSSRLETDATLLKTIV 239
Y + H + +MGERLT RVR +M SAIL NE+ WFD N + L++ L DATL+++ +
Sbjct: 4 YLLLHYFYTLMGERLTARVRLLMFSAILNNEVAWFDKDENNTGSLTAMLAADATLVRSAL 183
Query: 240 VDRSTILLQNVGLVVTALVIAFILNWRITLVVLATYPLIISGHIGEKLFMQGFGGNLSKA 299
DR + ++QNV L VTA VI F L+W++T VV+A PL+I I E+LF++GFGG+ A
Sbjct: 184 ADRLSTIVQNVALTVTAFVIGFTLSWKLTAVVVACLPLLIGASITEQLFLKGFGGDYGHA 363
Query: 300 YLKANMLAGEAVSNIRTVAAFCAEEKVIDLYADELVEPSKRSFKRGQIAGIFYGISQFFI 359
Y +A LA EA++NIRTVAAF A ++V +A EL +P+K++ RG I+G YGI+Q
Sbjct: 364 YSRATSLAREAIANIRTVAAFGAXDRVSTQFASELNKPNKQALLRGHISGFGYGITQLLA 543
Query: 360 FSSYGL 365
F SY L
Sbjct: 544 FCSYAL 561
>TC213339 similar to UP|Q9ZRG2 (Q9ZRG2) P-glycoprotein, partial (15%)
Length = 582
Score = 121 bits (303), Expect(2) = 2e-44
Identities = 58/86 (67%), Positives = 72/86 (83%)
Frame = +1
Query: 500 GKVMIDGKDIKKMNLKSLRKQIGLVQQEPALFATSIYKNILYGKEEASESEVIEAAKLAD 559
G+VMIDGKDI+K NLKSLR+ I +V QEP LFAT+IY+NI YG + AS++E+IEAA LA+
Sbjct: 325 GQVMIDGKDIRKYNLKSLRRHIAVVPQEPCLFATTIYENIAYGHDSASDAEIIEAATLAN 504
Query: 560 AHNFISALPEGYSTKAGDRGVLLSGG 585
AH FIS+LP+GY T G+RGV LSGG
Sbjct: 505 AHKFISSLPDGYKTFVGERGVQLSGG 582
Score = 77.4 bits (189), Expect(2) = 2e-44
Identities = 40/110 (36%), Positives = 69/110 (62%), Gaps = 3/110 (2%)
Frame = +3
Query: 365 LALWYGSVLLEKELASFKSIMKSFMVLIVTALAMGETLALAPDLLKGNQMVSSIFDMIDR 424
LA Y S L++ ++ F + ++ FMVL+V+A ETL LAPD +KG + S FD++DR
Sbjct: 3 LASGYASWLVKHGISDFSNTIRVFMVLMVSANGAAETLTLAPDFIKGGHAMRSAFDLLDR 182
Query: 425 KSGIIHDVGEELM---TVEGMIELKRINFIYPSRPNVVIFKDFNLIVPSG 471
++ I D + ++ G +ELK ++F YP+RP++ +F++ +L +G
Sbjct: 183 RTEIEPDDPDATPVPDSLRGEVELKHVDFSYPTRPDMSVFRNLSLRARAG 332
>AW308969 similar to GP|6671365|gb P-glycoprotein {Gossypium hirsutum},
partial (9%)
Length = 449
Score = 174 bits (441), Expect = 1e-43
Identities = 88/108 (81%), Positives = 101/108 (93%)
Frame = +3
Query: 369 YGSVLLEKELASFKSIMKSFMVLIVTALAMGETLALAPDLLKGNQMVSSIFDMIDRKSGI 428
YGSVL+EKELASFKSIMK+F VLIVTALAMGETLALAPDLLKGNQMV+S+F+++DRKSGI
Sbjct: 126 YGSVLMEKELASFKSIMKAFFVLIVTALAMGETLALAPDLLKGNQMVASVFEVMDRKSGI 305
Query: 429 IHDVGEELMTVEGMIELKRINFIYPSRPNVVIFKDFNLIVPSGKSLAL 476
+VGEEL TV+G IELKRINF YPSRP+V+IFKDFNL VP+GKS+AL
Sbjct: 306 SCEVGEELKTVDGTIELKRINFSYPSRPDVIIFKDFNLRVPAGKSVAL 449
>TC219347 similar to UP|Q9LF78 (Q9LF78) Mitochondrial half-ABC transporter,
partial (36%)
Length = 1085
Score = 172 bits (437), Expect = 3e-43
Identities = 89/177 (50%), Positives = 126/177 (70%)
Frame = +1
Query: 441 GMIELKRINFIYPSRPNVVIFKDFNLIVPSGKSLALVGHSGSGKSSIISLILRFYDPTSG 500
G I+ + ++F Y + ++ + +VP+GKS+A+VG SGSGKS+I+ L+ RF+DP SG
Sbjct: 46 GRIQFENVHFSYLTERKIL--DGISFVVPAGKSVAIVGTSGSGKSTILRLLFRFFDPHSG 219
Query: 501 KVMIDGKDIKKMNLKSLRKQIGLVQQEPALFATSIYKNILYGKEEASESEVIEAAKLADA 560
+ ID ++I+++ L+SLRK IG+V Q+ LF +I+ NI YG+ A+E EV EAA+ A
Sbjct: 220 SIKIDDQNIREVTLESLRKSIGVVPQDTVLFNDTIFHNIHYGRLSATEEEVYEAAQQAAI 399
Query: 561 HNFISALPEGYSTKAGDRGVLLSGGQKQRVAIARAILRNPKILLLDEATSALDVESE 617
HN I P+ YST G+RG+ LSGG+KQRVA+ARA L+ P ILL DEATSALD +E
Sbjct: 400 HNTIMKFPDKYSTVVGERGLKLSGGEKQRVALARAFLKAPAILLCDEATSALDSTTE 570
>TC207596 similar to UP|Q7GBY1 (Q7GBY1) Tonoplast ABC transporter IDI7,
partial (35%)
Length = 1076
Score = 164 bits (416), Expect = 8e-41
Identities = 85/148 (57%), Positives = 112/148 (75%), Gaps = 1/148 (0%)
Frame = +2
Query: 471 GKSLALVGHSGSGKSSIISLILRFYDPTSGKVMIDGKDIKKMNLKSLRKQIGLVQQEPAL 530
G +ALVG SG GKS+I +LI RFYDPT GK++++G + +++ K L ++I +V QEP L
Sbjct: 20 GSKVALVGPSGGGKSTIANLIERFYDPTKGKILLNGVPLVEISHKHLHRKISIVSQEPTL 199
Query: 531 FATSIYKNILYGKE-EASESEVIEAAKLADAHNFISALPEGYSTKAGDRGVLLSGGQKQR 589
F SI +NI YG + + ++ ++ AAK+A+AH FIS PE Y T G+RGV LSGGQKQR
Sbjct: 200 FNCSIEENIAYGFDGKVNDVDIENAAKMANAHEFISKFPEKYQTFVGERGVRLSGGQKQR 379
Query: 590 VAIARAILRNPKILLLDEATSALDVESE 617
+AIARA+L +PKILLLDEATSALD ESE
Sbjct: 380 IAIARALLMDPKILLLDEATSALDAESE 463
Score = 66.2 bits (160), Expect = 4e-11
Identities = 31/54 (57%), Positives = 42/54 (77%)
Frame = +2
Query: 1 MIGRTTVIVAHRLSTIKNADMIAVVEGGSVVETGNHEELISNPNSLYSSLVQGQ 54
M GRT +++AHRLST+K AD +AV+ G VVE GNHEEL+ N N +Y++LV+ Q
Sbjct: 494 MKGRTVLVIAHRLSTVKTADTVAVISDGQVVERGNHEELL-NKNGVYTALVKRQ 652
>AW507715 weakly similar to GP|9294227|db P-glycoprotein; multi-drug
resistance related; ABC transporter-like protein
{Arabidopsis thaliana}, partial (17%)
Length = 655
Score = 159 bits (403), Expect = 3e-39
Identities = 86/154 (55%), Positives = 110/154 (70%)
Frame = +2
Query: 466 LIVPSGKSLALVGHSGSGKSSIISLILRFYDPTSGKVMIDGKDIKKMNLKSLRKQIGLVQ 525
L + +G ALVG + SGK+S+I+LI FYDP + KVM+DGKDI K NLKS+R +IGL
Sbjct: 8 LRIRAGHIQALVGANSSGKTSVIALIDTFYDPIAVKVMVDGKDIMKSNLKSMRLKIGLAP 187
Query: 526 QEPALFATSIYKNILYGKEEASESEVIEAAKLADAHNFISALPEGYSTKAGDRGVLLSGG 585
EPALFA SI NI Y E A+E++VI AA+ A+ H F+S LP+GY+T G R V LS G
Sbjct: 188 LEPALFAASIVDNIAYATEGAAEADVIAAARAANVHGFVSGLPKGYNTPGGQRCVRLSRG 367
Query: 586 QKQRVAIARAILRNPKILLLDEATSALDVESERV 619
+AIARA+L++P IL+L E T+ALD ESE V
Sbjct: 368 LTLTIAIARAVLKDPTILVLYEVTTALDAESECV 469
>AW308906
Length = 445
Score = 154 bits (390), Expect = 9e-38
Identities = 79/147 (53%), Positives = 108/147 (72%), Gaps = 3/147 (2%)
Frame = +3
Query: 388 FMVLIVTALAMGETLALAPDLLKGNQMVSSIFDMIDRKSGIIHDVGEELM---TVEGMIE 444
FMVL+V+A ETL LAPD +KG + S FD++DR++ I D + ++ G +E
Sbjct: 3 FMVLMVSANGAAETLTLAPDFIKGGHAMRSAFDLLDRRTEIEPDDPDATPVPDSLRGEVE 182
Query: 445 LKRINFIYPSRPNVVIFKDFNLIVPSGKSLALVGHSGSGKSSIISLILRFYDPTSGKVMI 504
LK ++F YP+RP++ +F++ +L +GK+LALVG SG GKSS+I+LI RFYDPTSG+VMI
Sbjct: 183 LKHVDFSYPTRPDMSVFRNLSLRARAGKTLALVGPSGCGKSSVIALIQRFYDPTSGQVMI 362
Query: 505 DGKDIKKMNLKSLRKQIGLVQQEPALF 531
DGKDI+K NLKSLR+ I +V QEP LF
Sbjct: 363 DGKDIRKYNLKSLRRHIAVVPQEPCLF 443
>TC208899 similar to UP|Q9LHD1 (Q9LHD1) Multidrug resistance p-glycoprotein,
partial (14%)
Length = 983
Score = 153 bits (387), Expect = 2e-37
Identities = 81/121 (66%), Positives = 95/121 (77%), Gaps = 3/121 (2%)
Frame = +3
Query: 502 VMIDGKDIKKMNLKSLRKQIGLVQQEPALFATSIYKNILYGK---EEASESEVIEAAKLA 558
V IDG DIK NLKSLRK I LV QEP LF +I +NI YG+ E ESE+IEAA+ A
Sbjct: 6 VTIDGMDIKSYNLKSLRKHIALVSQEPTLFGGTIRENIAYGRCESERVDESEIIEAARAA 185
Query: 559 DAHNFISALPEGYSTKAGDRGVLLSGGQKQRVAIARAILRNPKILLLDEATSALDVESER 618
+AH+FI++L EGY T GD+GV LSGGQKQR+AIARAIL+NPK+LLLDEATSALD SE+
Sbjct: 186 NAHDFIASLKEGYETWCGDKGVQLSGGQKQRIAIARAILKNPKVLLLDEATSALDGPSEK 365
Query: 619 V 619
V
Sbjct: 366 V 368
Score = 53.5 bits (127), Expect = 3e-07
Identities = 30/55 (54%), Positives = 36/55 (64%), Gaps = 1/55 (1%)
Frame = +3
Query: 1 MIGRTTVIVAHRLSTIKNADMIAVVEGGSVVETGNHEELISNPN-SLYSSLVQGQ 54
M GRT V+VAHRLSTI N D+I V+E G VVE G H L++ + Y SLV Q
Sbjct: 393 MRGRTGVVVAHRLSTIHNCDVIGVLEKGRVVEIGTHSSLLAKGSCGAYYSLVSLQ 557
>TC234247 similar to GB|CAD59589.1|27368863|OSA535067 MDR-like ABC
transporter {Oryza sativa (japonica cultivar-group);} ,
partial (11%)
Length = 422
Score = 89.7 bits (221), Expect(2) = 1e-30
Identities = 42/79 (53%), Positives = 59/79 (74%)
Frame = +1
Query: 436 LMTVEGMIELKRINFIYPSRPNVVIFKDFNLIVPSGKSLALVGHSGSGKSSIISLILRFY 495
L + G IEL+ + F YP+RP+ +IF F+L +PSG + ALVG SGSGKS+++SLI RFY
Sbjct: 1 LEDIRGDIELREVCFSYPTRPDELIFNGFSLSIPSGTTAALVGQSGSGKSTVVSLIERFY 180
Query: 496 DPTSGKVMIDGKDIKKMNL 514
DP SG V+IDG ++++ L
Sbjct: 181 DPQSGAVLIDGINLREFQL 237
Score = 62.4 bits (150), Expect(2) = 1e-30
Identities = 30/57 (52%), Positives = 41/57 (71%)
Frame = +2
Query: 517 LRKQIGLVQQEPALFATSIYKNILYGKEEASESEVIEAAKLADAHNFISALPEGYST 573
+R++IGLV QEP LF SI +NI YGK+ A++ E+ AA+LA+A FI LP+G T
Sbjct: 245 IRQKIGLVSQEPVLFTCSIKENIAYGKDGATDEEIRAAAELANAAKFIDKLPQGLDT 415
>TC205367 similar to UP|O24525 (O24525) Glutathione-conjugate transporter
AtMRP4, partial (23%)
Length = 1548
Score = 113 bits (283), Expect = 2e-25
Identities = 65/185 (35%), Positives = 108/185 (58%), Gaps = 5/185 (2%)
Frame = +2
Query: 440 EGMIELKRINFIYPSRPNV-VIFKDFNLIVPSGKSLALVGHSGSGKSSIISLILRFYDPT 498
EG +++K + Y RPN ++ K L + G+ + +VG +GSGKS++I + R +PT
Sbjct: 332 EGHVDIKDLQVRY--RPNTPLVLKGITLSINGGEKIGVVGRTGSGKSTLIQVFFRLVEPT 505
Query: 499 SGKVMIDGKDIKKMNLKSLRKQIGLVQQEPALFATSIYKNI----LYGKEEASESEVIEA 554
GK++IDG DI + L LR + G++ QEP LF ++ NI Y EE +S +E
Sbjct: 506 GGKIIIDGIDISALGLHDLRSRFGIIPQEPVLFEGTVRSNIDPTGQYTDEEIWKS--LER 679
Query: 555 AKLADAHNFISALPEGYSTKAGDRGVLLSGGQKQRVAIARAILRNPKILLLDEATSALDV 614
+L DA +++ PE T D G S GQ+Q + + R +L+ ++L +DEAT+++D
Sbjct: 680 CQLKDA---VASKPEKLDTSVVDNGDNWSVGQRQLLCLGRVMLKQSRLLFMDEATASVDS 850
Query: 615 ESERV 619
+++ V
Sbjct: 851 QTDAV 865
>AW705127 weakly similar to GP|27359084|gb ABC-type bacteriocin/lantibiotic
exporter {Vibrio vulnificus CMCP6}, partial (16%)
Length = 428
Score = 108 bits (270), Expect = 7e-24
Identities = 54/128 (42%), Positives = 83/128 (64%)
Frame = +2
Query: 490 LILRFYDPTSGKVMIDGKDIKKMNLKSLRKQIGLVQQEPALFATSIYKNILYGKEEASES 549
L+ Y PT+G V IDG D+++++ R+ IG+V+QEP LF ++ +N+L G+E A
Sbjct: 5 LLAGLYQPTAGAVEIDGLDLRQLDPADFRRHIGVVEQEPRLFRGTVRENLLLGRE-AGAD 181
Query: 550 EVIEAAKLADAHNFISALPEGYSTKAGDRGVLLSGGQKQRVAIARAILRNPKILLLDEAT 609
+ E +L ++A P G G+ G +LSGGQ+Q VA+ARA+L P+ILL+DE T
Sbjct: 182 RLAEVLRLTGLDRVVAAHPMGLDMPVGEGGAMLSGGQRQLVALARALLLRPQILLMDEPT 361
Query: 610 SALDVESE 617
SA+D ++E
Sbjct: 362 SAMDAQTE 385
>TC229512 similar to UP|Q9SDM5 (Q9SDM5) P-glycoprotein, partial (11%)
Length = 633
Score = 106 bits (264), Expect = 3e-23
Identities = 54/69 (78%), Positives = 60/69 (86%)
Frame = +1
Query: 551 VIEAAKLADAHNFISALPEGYSTKAGDRGVLLSGGQKQRVAIARAILRNPKILLLDEATS 610
VIEA KLA+AHNFIS L EG TK G RGV L GGQ+QRVAIARA+L+NP+ILLLDEATS
Sbjct: 1 VIEAVKLANAHNFISGLXEGXXTKVGXRGVQLXGGQRQRVAIARAVLKNPEILLLDEATS 180
Query: 611 ALDVESERV 619
ALDVESER+
Sbjct: 181 ALDVESERI 207
Score = 61.6 bits (148), Expect = 1e-09
Identities = 29/54 (53%), Positives = 37/54 (67%)
Frame = +1
Query: 1 MIGRTTVIVAHRLSTIKNADMIAVVEGGSVVETGNHEELISNPNSLYSSLVQGQ 54
M RTTV+VAHRLSTI+NAD I+V++ G +++ G H LI N N Y LV Q
Sbjct: 232 MQNRTTVMVAHRLSTIRNADQISVLQDGKIIDQGTHSSLIENKNGAYYKLVNLQ 393
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.136 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,188,459
Number of Sequences: 63676
Number of extensions: 268525
Number of successful extensions: 1602
Number of sequences better than 10.0: 128
Number of HSP's better than 10.0 without gapping: 1539
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1576
length of query: 619
length of database: 12,639,632
effective HSP length: 103
effective length of query: 516
effective length of database: 6,081,004
effective search space: 3137798064
effective search space used: 3137798064
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC149574.1


