

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
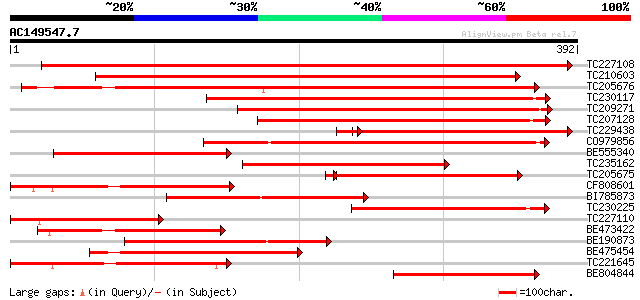
Query= AC149547.7 - phase: 0
(392 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC227108 similar to UP|Q9M3V0 (Q9M3V0) Protein phosphatase 2C (P... 594 e-170
TC210603 similar to UP|Q9LHJ9 (Q9LHJ9) Protein phosphatase 2C (A... 405 e-113
TC205676 similar to GB|AAL87371.1|19548015|AY081718 AT4g33920/F1... 390 e-109
TC230117 similar to UP|Q9LSN8 (Q9LSN8) Protein phosphatase 2C-li... 313 9e-86
TC209271 similar to UP|Q9LHJ9 (Q9LHJ9) Protein phosphatase 2C (A... 285 2e-77
TC207128 similar to UP|Q9LSN8 (Q9LSN8) Protein phosphatase 2C-li... 275 3e-74
TC229438 similar to UP|Q9ZSQ7 (Q9ZSQ7) Protein phosphatase 2C ho... 258 7e-74
CO979856 266 1e-71
BE555340 similar to GP|20340237|gb protein phosphatase 2c-like p... 243 1e-64
TC235162 similar to UP|O82479 (O82479) Protein phosphatase-2c (F... 201 5e-52
TC205675 similar to GB|AAL87371.1|19548015|AY081718 AT4g33920/F1... 167 7e-42
CF808601 164 6e-41
BI785873 162 3e-40
TC230225 similar to UP|Q9FG32 (Q9FG32) Protein phosphatase 2C-li... 154 7e-38
TC227110 similar to UP|Q9M3V0 (Q9M3V0) Protein phosphatase 2C (P... 147 6e-36
BE473422 similar to GP|13937198|gb| AT4g33920/F17I5_110 {Arabido... 139 2e-33
BE190873 weakly similar to GP|21536936|gb| protein phosphatase 2... 135 2e-32
BE475454 130 1e-30
TC221645 similar to UP|Q9LSN8 (Q9LSN8) Protein phosphatase 2C-li... 124 7e-29
BE804844 similar to GP|13937198|gb| AT4g33920/F17I5_110 {Arabido... 122 4e-28
>TC227108 similar to UP|Q9M3V0 (Q9M3V0) Protein phosphatase 2C (PP2C) ,
partial (94%)
Length = 1387
Score = 594 bits (1532), Expect = e-170
Identities = 289/367 (78%), Positives = 328/367 (88%)
Frame = +2
Query: 23 SDVCGKKEGLLWYKDAGQHLFGDYSMAVVQANNLLEDQSQIESGPLSFLDTGPYGTFVGV 82
SD G+++GLLWYKD+GQHL G++SMAV+QANNLLEDQSQIESG LS ++GPYGTF+GV
Sbjct: 20 SDAGGRQDGLLWYKDSGQHLSGEFSMAVIQANNLLEDQSQIESGCLSSNESGPYGTFIGV 199
Query: 83 YDGHGGPETSRFICDHLFQHLKRFATEHKSMSVEVIRKAYQATEEGFLGVVTKHWPMNPQ 142
YDGHGGPETSRFI DHLF HLKRF +E +SMSV+VIRKA QATEEGF+ VV + + ++PQ
Sbjct: 200 YDGHGGPETSRFINDHLFHHLKRFTSEQQSMSVDVIRKALQATEEGFISVVARQFSLSPQ 379
Query: 143 IAAVGSCCLVGVICGGSLYIANLGDSRAVLGRAVRATGEVLAIQLSPEHNVAIESVRQEM 202
IAAVGSCCLVGVIC G+LYIANLGDSRAVLGRAV+ATGEVLA+QLS EHN +IE+VRQE+
Sbjct: 380 IAAVGSCCLVGVICNGTLYIANLGDSRAVLGRAVKATGEVLAMQLSAEHNASIETVRQEL 559
Query: 203 HSLHPDDPKIVVLKHNVWRVKGLIQISRSIGDVYLKKAEFNREPLYAKFRLRETFKTPIL 262
H+ HPDDP IVVLKHNVWRVKGLIQ+SRSIGDVYLKKAEFNREPLYAKFRLRE +K PIL
Sbjct: 560 HASHPDDPNIVVLKHNVWRVKGLIQVSRSIGDVYLKKAEFNREPLYAKFRLREPYKMPIL 739
Query: 263 SSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAVDIVQNHPHSGSARKLIKVALLEAAK 322
SS+PSISVH LQ HDQF+IFASDGLWEHLSNQ+AVDIVQN P SGSAR+L+K AL EAAK
Sbjct: 740 SSEPSISVHHLQPHDQFIIFASDGLWEHLSNQEAVDIVQNSPRSGSARRLVKAALQEAAK 919
Query: 323 KREMRYSDLKKIDRGVRRHFHDDITVVVIFLDSNLVSRASTVTGPPVSLRGAGVPLPSRS 382
KREMRYSDLKKIDRGVRRHFHDD TV+V++LDSNLVSRASTV P +S+RG G+ LP +
Sbjct: 920 KREMRYSDLKKIDRGVRRHFHDDTTVIVVYLDSNLVSRASTVKFPGISVRGGGINLPHNT 1099
Query: 383 LAPMELP 389
LAP P
Sbjct: 1100LAPCTTP 1120
>TC210603 similar to UP|Q9LHJ9 (Q9LHJ9) Protein phosphatase 2C (At3g12620),
partial (75%)
Length = 883
Score = 405 bits (1041), Expect = e-113
Identities = 197/294 (67%), Positives = 236/294 (80%)
Frame = +2
Query: 60 QSQIESGPLSFLDTGPYGTFVGVYDGHGGPETSRFICDHLFQHLKRFATEHKSMSVEVIR 119
QSQ+ESGPLS + P GTFVG+YDGHGGPE +RF+ D LF ++K+F +E+ MS +VI
Sbjct: 2 QSQLESGPLSLTEGNPQGTFVGIYDGHGGPEAARFVNDRLFNNIKKFTSENNGMSADVIN 181
Query: 120 KAYQATEEGFLGVVTKHWPMNPQIAAVGSCCLVGVICGGSLYIANLGDSRAVLGRAVRAT 179
KA+ ATEE FL +V K W P IA+VGSCCL+G+IC G LYIAN GDSRAVLGR A
Sbjct: 182 KAFLATEEEFLSLVEKLWLHKPPIASVGSCCLIGIICSGELYIANAGDSRAVLGRLDEAM 361
Query: 180 GEVLAIQLSPEHNVAIESVRQEMHSLHPDDPKIVVLKHNVWRVKGLIQISRSIGDVYLKK 239
E+ IQLS EHN + SVR+E+HSLHP+DP+IVV+KH VWRVKGLIQISRSIGD YLKK
Sbjct: 362 KEIKDIQLSVEHNASHASVREELHSLHPNDPQIVVMKHQVWRVKGLIQISRSIGDAYLKK 541
Query: 240 AEFNREPLYAKFRLRETFKTPILSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAVDI 299
AEFN+ PL KFRL E F PIL ++P+I V +L DQFLI ASDGLWE LSNQ+AV+I
Sbjct: 542 AEFNKAPLLPKFRLSEPFDQPILKAEPAILVQKLCPQDQFLILASDGLWERLSNQEAVNI 721
Query: 300 VQNHPHSGSARKLIKVALLEAAKKREMRYSDLKKIDRGVRRHFHDDITVVVIFL 353
VQ+ P +G+A+KL+K AL EAAKKREMRYSDL+KIDRGVRRHFHDDITV+V++L
Sbjct: 722 VQSCPRNGAAKKLVKTALCEAAKKREMRYSDLRKIDRGVRRHFHDDITVIVLYL 883
>TC205676 similar to GB|AAL87371.1|19548015|AY081718 AT4g33920/F17I5_110
{Arabidopsis thaliana;} , partial (81%)
Length = 1793
Score = 390 bits (1003), Expect = e-109
Identities = 201/360 (55%), Positives = 253/360 (69%), Gaps = 2/360 (0%)
Frame = +2
Query: 9 LTACWRRRSSDRKSSDVCGKKEGLLWYKDAGQHLFGDYSMAVVQANNLLEDQSQIESGPL 68
L C+RRR +D GLLW+ D H GD+S+AV QAN LEDQSQ+
Sbjct: 155 LDCCFRRRRAD-----------GLLWHTDLKPHASGDFSIAVAQANYCLEDQSQVF---- 289
Query: 69 SFLDTGPYGTFVGVYDGHGGPETSRFICDHLFQHLKRFATEHKSMSVEVIRKAYQATEEG 128
T PY T+VGVYDGHGGPE SRF+ LF +L +FATE +SV+VI+KA+ ATEE
Sbjct: 290 ----TSPYATYVGVYDGHGGPEASRFVNKRLFPYLHKFATEQGGLSVDVIKKAFSATEEE 457
Query: 129 FLGVVTKHWPMNPQIAAVGSCCLVGVICGGSLYIANLGDSRAVLGR--AVRATGEVLAIQ 186
FL +V P++PQIA+VGSCCL G I LY+ANLGDSRAVLGR VR V+A +
Sbjct: 458 FLHLVKLSLPISPQIASVGSCCLFGAISNNVLYVANLGDSRAVLGRRDTVRKNSPVVAQR 637
Query: 187 LSPEHNVAIESVRQEMHSLHPDDPKIVVLKHNVWRVKGLIQISRSIGDVYLKKAEFNREP 246
LS +HNVA E VR+E+ +LHPDD IVV VWR+KG+IQ+SRSIGDVYLKK +F R+P
Sbjct: 638 LSTDHNVADEEVRKEVEALHPDDSHIVVYNRGVWRIKGIIQVSRSIGDVYLKKPDFYRDP 817
Query: 247 LYAKFRLRETFKTPILSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAVDIVQNHPHS 306
++ +F K P+++++PSI + EL+ D FLIFASDGLWE LS++ AV IV HP +
Sbjct: 818 VFQQFGNPIPLKRPVMTAEPSIIIRELESQDLFLIFASDGLWEQLSDEAAVQIVFKHPRA 997
Query: 307 GSARKLIKVALLEAAKKREMRYSDLKKIDRGVRRHFHDDITVVVIFLDSNLVSRASTVTG 366
G A++L++ AL EAAKKREMRY D+KKID+G+RRHFHDDITVVVI+LD + S T G
Sbjct: 998 GIAKRLVRAALHEAAKKREMRYDDIKKIDKGIRRHFHDDITVVVIYLDHHAGSSKQTAVG 1177
>TC230117 similar to UP|Q9LSN8 (Q9LSN8) Protein phosphatase 2C-like protein,
partial (59%)
Length = 888
Score = 313 bits (802), Expect = 9e-86
Identities = 155/238 (65%), Positives = 188/238 (78%)
Frame = +3
Query: 137 WPMNPQIAAVGSCCLVGVICGGSLYIANLGDSRAVLGRAVRATGEVLAIQLSPEHNVAIE 196
W PQIA G+CCLVGVIC +L++A+LGDSRAVLGR V TG + AIQLS EHN E
Sbjct: 6 WSSRPQIATTGTCCLVGVICRQTLFVASLGDSRAVLGRRVGNTGGMAAIQLSTEHNANFE 185
Query: 197 SVRQEMHSLHPDDPKIVVLKHNVWRVKGLIQISRSIGDVYLKKAEFNREPLYAKFRLRET 256
+V E+ LHP+DP+IVVLKH VWRVKG+IQ+SRSIGDVY+K A+FNREP+ AKFRL E
Sbjct: 186 AVMFELKELHPNDPQIVVLKHGVWRVKGIIQVSRSIGDVYMKHAQFNREPINAKFRLPEP 365
Query: 257 FKTPILSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAVDIVQNHPHSGSARKLIKVA 316
P LS++P+I H LQ +D FLIFASDGLWEHLSN AVDIV + P +GSA+KL+K A
Sbjct: 366 MNMPFLSANPTILSHPLQPNDSFLIFASDGLWEHLSNDQAVDIVHSSPCAGSAKKLVKAA 545
Query: 317 LLEAAKKREMRYSDLKKIDRGVRRHFHDDITVVVIFLDSNLVSRASTVTGPPVSLRGA 374
L EAA+KREMRYSDL KID+ VRRHFHDDITV+V+FL+ NL+SR + V P+++R A
Sbjct: 546 LQEAARKREMRYSDLYKIDKKVRRHFHDDITVIVLFLNHNLISRGA-VLNTPLTIRSA 716
>TC209271 similar to UP|Q9LHJ9 (Q9LHJ9) Protein phosphatase 2C (At3g12620),
partial (49%)
Length = 720
Score = 285 bits (730), Expect = 2e-77
Identities = 141/218 (64%), Positives = 173/218 (78%)
Frame = +1
Query: 158 GSLYIANLGDSRAVLGRAVRATGEVLAIQLSPEHNVAIESVRQEMHSLHPDDPKIVVLKH 217
G LY+AN GDSRAVLGR RAT E IQLS EHNV I++ R E+ + HP DP+IVV+KH
Sbjct: 10 GMLYVANAGDSRAVLGRVERATRETTTIQLSAEHNVNIQTERDEVRTKHPYDPQIVVMKH 189
Query: 218 NVWRVKGLIQISRSIGDVYLKKAEFNREPLYAKFRLRETFKTPILSSDPSISVHELQEHD 277
NVWRVKG+IQ+SRSIGD YLKK EFNREPL KFRL E F PILS +P+ISVH+L+ D
Sbjct: 190 NVWRVKGIIQVSRSIGDAYLKKDEFNREPLPNKFRLSEPFSKPILSYEPAISVHKLRPED 369
Query: 278 QFLIFASDGLWEHLSNQDAVDIVQNHPHSGSARKLIKVALLEAAKKREMRYSDLKKIDRG 337
QF+IFASDGLWE LSNQ+ V+IV N P +G AR+L+K AL AA+KREMR SDL+KI++G
Sbjct: 370 QFIIFASDGLWEQLSNQEVVNIVSNSPRNGIARRLVKAALRVAARKREMRVSDLQKIEQG 549
Query: 338 VRRHFHDDITVVVIFLDSNLVSRASTVTGPPVSLRGAG 375
VRRHFHDDITV+V+FL+ L+ +S + P+S++G G
Sbjct: 550 VRRHFHDDITVIVVFLNHKLIDNSSLLAS-PLSIKGGG 660
>TC207128 similar to UP|Q9LSN8 (Q9LSN8) Protein phosphatase 2C-like protein,
partial (49%)
Length = 962
Score = 275 bits (703), Expect = 3e-74
Identities = 137/203 (67%), Positives = 167/203 (81%)
Frame = +2
Query: 172 LGRAVRATGEVLAIQLSPEHNVAIESVRQEMHSLHPDDPKIVVLKHNVWRVKGLIQISRS 231
LG+ V TG + AIQLS EHN +E+VRQE+ LHP DP+IVVLKH VWRVKG+IQ+SRS
Sbjct: 2 LGKKVGNTGGMAAIQLSTEHNANLEAVRQELKELHPHDPQIVVLKHGVWRVKGIIQVSRS 181
Query: 232 IGDVYLKKAEFNREPLYAKFRLRETFKTPILSSDPSISVHELQEHDQFLIFASDGLWEHL 291
IGDVYLK A+FNREPL AKFRL E PILS++P+I H LQ +D FLIFASDGLWEHL
Sbjct: 182 IGDVYLKHAQFNREPLNAKFRLPEPMNMPILSANPTILSHALQPNDSFLIFASDGLWEHL 361
Query: 292 SNQDAVDIVQNHPHSGSARKLIKVALLEAAKKREMRYSDLKKIDRGVRRHFHDDITVVVI 351
SN+ AVDIV ++PH+GSA++LIK AL EAA+KREMRYSDL+KID+ VRRHFHDDI+V+V+
Sbjct: 362 SNEKAVDIVNSNPHAGSAKRLIKAALHEAARKREMRYSDLRKIDKKVRRHFHDDISVIVL 541
Query: 352 FLDSNLVSRASTVTGPPVSLRGA 374
FL+ +L+SR TV P +S+R A
Sbjct: 542 FLNHDLISR-GTVLDPTLSIRSA 607
>TC229438 similar to UP|Q9ZSQ7 (Q9ZSQ7) Protein phosphatase 2C homolog,
partial (42%)
Length = 832
Score = 258 bits (659), Expect(2) = 7e-74
Identities = 127/152 (83%), Positives = 138/152 (90%)
Frame = +3
Query: 238 KKAEFNREPLYAKFRLRETFKTPILSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAV 297
K+ REPLYAKFR+RE FK PILSSDPSISVHELQ+HDQFLIFASDGLWEHLSNQDAV
Sbjct: 282 KRLNSTREPLYAKFRVREGFKRPILSSDPSISVHELQQHDQFLIFASDGLWEHLSNQDAV 461
Query: 298 DIVQNHPHSGSARKLIKVALLEAAKKREMRYSDLKKIDRGVRRHFHDDITVVVIFLDSNL 357
DIVQN+PH+G AR+LIK AL EAAKKREMRYSDLKKIDRGVRRHFHDDITVVV+FLDSNL
Sbjct: 462 DIVQNNPHNGIARRLIKAALQEAAKKREMRYSDLKKIDRGVRRHFHDDITVVVVFLDSNL 641
Query: 358 VSRASTVTGPPVSLRGAGVPLPSRSLAPMELP 389
VSRAS+V GPP+S+RG GVPLPSR+LAP P
Sbjct: 642 VSRASSVRGPPLSVRGGGVPLPSRTLAPCAAP 737
Score = 37.7 bits (86), Expect(2) = 7e-74
Identities = 17/18 (94%), Positives = 18/18 (99%)
Frame = +2
Query: 227 QISRSIGDVYLKKAEFNR 244
QISRSIGDVYLKKAEFN+
Sbjct: 248 QISRSIGDVYLKKAEFNK 301
>CO979856
Length = 861
Score = 266 bits (680), Expect = 1e-71
Identities = 129/239 (53%), Positives = 179/239 (73%)
Frame = -2
Query: 135 KHWPMNPQIAAVGSCCLVGVICGGSLYIANLGDSRAVLGRAVRATGEVLAIQLSPEHNVA 194
+ + + P IA++GSCCLVGVI G+LYIANLGDSRAV+G R+ +++A QL+ EHN
Sbjct: 860 RSYMIKPLIASIGSCCLVGVIWKGTLYIANLGDSRAVVGSLGRSN-KIIAEQLTREHNAC 684
Query: 195 IESVRQEMHSLHPDDPKIVVLKHNVWRVKGLIQISRSIGDVYLKKAEFNREPLYAKFRLR 254
E +RQE+ SLHP D +IVV+ WRVKG+IQ+SRSIGD YLK +F+ +P + +F +
Sbjct: 683 REEIRQELRSLHPQDSQIVVMNRGTWRVKGIIQVSRSIGDAYLKWPQFSLDPSFPRFHMP 504
Query: 255 ETFKTPILSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDAVDIVQNHPHSGSARKLIK 314
E P+L+++PS+ LQ HD+FLIFASDGLWE+++NQ A +IVQ +P +G ARKL+K
Sbjct: 503 EPITQPVLTAEPSLCSRVLQPHDKFLIFASDGLWEYMTNQQAAEIVQKNPRNGVARKLVK 324
Query: 315 VALLEAAKKREMRYSDLKKIDRGVRRHFHDDITVVVIFLDSNLVSRASTVTGPPVSLRG 373
AL EAA KR+M+Y +L+KI++G RR FHDDITV+V+F+D L+ + TV P +S+RG
Sbjct: 323 AALKEAANKRKMKYKELQKIEKGNRRIFHDDITVIVVFIDHELLGKKITV--PELSIRG 153
>BE555340 similar to GP|20340237|gb protein phosphatase 2c-like protein
{Thellungiella halophila}, partial (32%)
Length = 374
Score = 243 bits (620), Expect = 1e-64
Identities = 114/123 (92%), Positives = 118/123 (95%)
Frame = +2
Query: 31 GLLWYKDAGQHLFGDYSMAVVQANNLLEDQSQIESGPLSFLDTGPYGTFVGVYDGHGGPE 90
GLLWYKDAGQHLFG+YSMAVVQANNLLEDQSQIESGPLS LDTGPYGTFVGVYDGHGGPE
Sbjct: 5 GLLWYKDAGQHLFGEYSMAVVQANNLLEDQSQIESGPLSLLDTGPYGTFVGVYDGHGGPE 184
Query: 91 TSRFICDHLFQHLKRFATEHKSMSVEVIRKAYQATEEGFLGVVTKHWPMNPQIAAVGSCC 150
TSR++CDHLFQHLKRFA+E KSMS EVIRKAYQATEEGFL VVTK WPMNPQIAAVGSCC
Sbjct: 185 TSRYVCDHLFQHLKRFASEQKSMSEEVIRKAYQATEEGFLSVVTKQWPMNPQIAAVGSCC 364
Query: 151 LVG 153
LVG
Sbjct: 365 LVG 373
>TC235162 similar to UP|O82479 (O82479) Protein phosphatase-2c (Fragment),
partial (47%)
Length = 436
Score = 201 bits (511), Expect = 5e-52
Identities = 103/143 (72%), Positives = 114/143 (79%)
Frame = +1
Query: 162 IANLGDSRAVLGRAVRATGEVLAIQLSPEHNVAIESVRQEMHSLHPDDPKIVVLKHNVWR 221
+AN GDSR VLGR RAT E AIQLS EHNV ESVR E+ S HP D +IVVL+ NVWR
Sbjct: 1 VANSGDSRVVLGRLERATRETEAIQLSTEHNVNQESVRDELRSKHPFDSQIVVLRQNVWR 180
Query: 222 VKGLIQISRSIGDVYLKKAEFNREPLYAKFRLRETFKTPILSSDPSISVHELQEHDQFLI 281
VKGLIQ+SRSIGD YLKKAEFNR+PL AK+RL ETF PILS DPS S H L DQFLI
Sbjct: 181 VKGLIQVSRSIGDAYLKKAEFNRDPLPAKYRLAETFFRPILSCDPSTSSHTLHPDDQFLI 360
Query: 282 FASDGLWEHLSNQDAVDIVQNHP 304
FASDGLWEHL+NQ+AV+IV N+P
Sbjct: 361 FASDGLWEHLTNQEAVNIVSNNP 429
>TC205675 similar to GB|AAL87371.1|19548015|AY081718 AT4g33920/F17I5_110
{Arabidopsis thaliana;} , partial (36%)
Length = 889
Score = 167 bits (422), Expect(2) = 7e-42
Identities = 78/128 (60%), Positives = 103/128 (79%)
Frame = +2
Query: 227 QISRSIGDVYLKKAEFNREPLYAKFRLRETFKTPILSSDPSISVHELQEHDQFLIFASDG 286
++SRSIGDVYLKK +F R+P++ +F K P+++++PSI + EL+ D FLIFASDG
Sbjct: 107 KVSRSIGDVYLKKPDFYRDPVFQQFGNPIPLKRPVMTAEPSIIIRELESQDLFLIFASDG 286
Query: 287 LWEHLSNQDAVDIVQNHPHSGSARKLIKVALLEAAKKREMRYSDLKKIDRGVRRHFHDDI 346
LWE LS++ AV IV HP +G A++L++ AL EAAKKREMRY D+KKID+G+RRHFHDDI
Sbjct: 287 LWEQLSDEAAVQIVFKHPRAGIAKRLVRAALHEAAKKREMRYDDIKKIDKGIRRHFHDDI 466
Query: 347 TVVVIFLD 354
TVVVI+LD
Sbjct: 467 TVVVIYLD 490
Score = 21.9 bits (45), Expect(2) = 7e-42
Identities = 7/9 (77%), Positives = 9/9 (99%)
Frame = +3
Query: 219 VWRVKGLIQ 227
VWR+KG+IQ
Sbjct: 3 VWRIKGIIQ 29
>CF808601
Length = 564
Score = 164 bits (415), Expect = 6e-41
Identities = 82/161 (50%), Positives = 113/161 (69%), Gaps = 6/161 (3%)
Frame = +1
Query: 1 MLSTLMDFLTACWRR--RSSDRKSS-DVCGK--KEGLLWYKDAGQHLFGDYSMAVVQANN 55
ML LM+ + CW+ R +DR S V G+ K+GLLW++D G++ GD+SMAVVQAN
Sbjct: 106 MLQALMNLFSLCWKPFGRDADRIDSIGVIGREGKDGLLWFRDFGKYGSGDFSMAVVQANQ 285
Query: 56 LLEDQSQIESGPLSFLDTGPYGTFVGVYDGHGGPETSRFICDHLFQHLKRFATEHKS-MS 114
+LEDQSQIESGPL GTFVG+YDGHGGP+ SR++CDHLF+H + + E + ++
Sbjct: 286 VLEDQSQIESGPL--------GTFVGIYDGHGGPDASRYVCDHLFRHFQAISAESRGVVT 441
Query: 115 VEVIRKAYQATEEGFLGVVTKHWPMNPQIAAVGSCCLVGVI 155
E I +A++ TEEG++ +V+ W P IA+ G+CCLVGVI
Sbjct: 442 TETIERAFRQTEEGYMALVSGSWNARPHIASAGTCCLVGVI 564
>BI785873
Length = 421
Score = 162 bits (409), Expect = 3e-40
Identities = 78/140 (55%), Positives = 109/140 (77%)
Frame = +2
Query: 109 EHKSMSVEVIRKAYQATEEGFLGVVTKHWPMNPQIAAVGSCCLVGVICGGSLYIANLGDS 168
E+ SMS ++IR A ATE+GFL +V + + + P IAA+GSCCLVGV+ G+LYIANLGDS
Sbjct: 2 ENGSMSEDIIRSAVSATEDGFLTLVRRSYGIKPLIAAMGSCCLVGVVWKGTLYIANLGDS 181
Query: 169 RAVLGRAVRATGEVLAIQLSPEHNVAIESVRQEMHSLHPDDPKIVVLKHNVWRVKGLIQI 228
RAV+G +V + +++A QL+ EHN + E VR+E+ SLHP+D +IVV+K WR+KG+IQ+
Sbjct: 182 RAVIG-SVGRSNKIIAEQLTKEHNASKEEVRRELRSLHPEDSQIVVMKQGTWRIKGIIQV 358
Query: 229 SRSIGDVYLKKAEFNREPLY 248
SRSIGD YLK+ EF+ +P +
Sbjct: 359 SRSIGDAYLKRPEFSFDPSF 418
>TC230225 similar to UP|Q9FG32 (Q9FG32) Protein phosphatase 2C-like, partial
(39%)
Length = 813
Score = 154 bits (389), Expect = 7e-38
Identities = 74/137 (54%), Positives = 102/137 (74%)
Frame = +2
Query: 237 LKKAEFNREPLYAKFRLRETFKTPILSSDPSISVHELQEHDQFLIFASDGLWEHLSNQDA 296
L EF+ +P + +F L E + P+L+++PSI L+ +D+F+IFASDGLWEHL+NQ+A
Sbjct: 29 LXXPEFSXDPSFPRFHLPEPIRRPVLTAEPSICSRVLKPNDKFIIFASDGLWEHLTNQEA 208
Query: 297 VDIVQNHPHSGSARKLIKVALLEAAKKREMRYSDLKKIDRGVRRHFHDDITVVVIFLDSN 356
+IV N+P G AR+L+K AL EAA+KREMRY DL+KI +G+RR FHDDITVVV+F+D
Sbjct: 209 AEIVHNNPRIGIARRLLKAALNEAARKREMRYKDLQKIGKGIRRFFHDDITVVVVFIDHE 388
Query: 357 LVSRASTVTGPPVSLRG 373
L R VT P +S++G
Sbjct: 389 L--RGKNVTVPDLSIKG 433
>TC227110 similar to UP|Q9M3V0 (Q9M3V0) Protein phosphatase 2C (PP2C) ,
partial (27%)
Length = 767
Score = 147 bits (372), Expect = 6e-36
Identities = 72/108 (66%), Positives = 87/108 (79%), Gaps = 2/108 (1%)
Frame = +3
Query: 1 MLSTLMDFLTACWRRRSSD--RKSSDVCGKKEGLLWYKDAGQHLFGDYSMAVVQANNLLE 58
MLS LM+ L AC+R S R SD G+++GLLWYKD+GQHL GD+SMAV+QANNLLE
Sbjct: 444 MLSGLMNLLRACFRPGSDGFTRAGSDAGGRQDGLLWYKDSGQHLNGDFSMAVIQANNLLE 623
Query: 59 DQSQIESGPLSFLDTGPYGTFVGVYDGHGGPETSRFICDHLFQHLKRF 106
DQSQIESG LS ++ PYGTF+ VYDGHGGP+TS+FI +HLF HLK+F
Sbjct: 624 DQSQIESGCLSSNESSPYGTFISVYDGHGGPKTSQFINNHLFHHLKKF 767
>BE473422 similar to GP|13937198|gb| AT4g33920/F17I5_110 {Arabidopsis
thaliana}, partial (24%)
Length = 395
Score = 139 bits (350), Expect = 2e-33
Identities = 73/134 (54%), Positives = 90/134 (66%), Gaps = 4/134 (2%)
Frame = +2
Query: 20 RKSSDVC----GKKEGLLWYKDAGQHLFGDYSMAVVQANNLLEDQSQIESGPLSFLDTGP 75
RK +C G + LLW+ D H G+YS+AVVQAN+ LEDQ+Q+ T P
Sbjct: 17 RKPLKMCFGGGGNDDDLLWHTDLKPHASGNYSIAVVQANSSLEDQAQVF--------TSP 172
Query: 76 YGTFVGVYDGHGGPETSRFICDHLFQHLKRFATEHKSMSVEVIRKAYQATEEGFLGVVTK 135
TFVGVYDGHGGPE SRFI +HLF L++FATE +S EVI+KA++ATEE FL VV +
Sbjct: 173 SATFVGVYDGHGGPEASRFITNHLFSFLRKFATEEGDLSEEVIKKAFEATEEEFLRVVRE 352
Query: 136 HWPMNPQIAAVGSC 149
W PQIA+VGSC
Sbjct: 353 SWIARPQIASVGSC 394
>BE190873 weakly similar to GP|21536936|gb| protein phosphatase 2C-like
{Arabidopsis thaliana}, partial (24%)
Length = 427
Score = 135 bits (341), Expect = 2e-32
Identities = 67/143 (46%), Positives = 95/143 (65%)
Frame = +2
Query: 80 VGVYDGHGGPETSRFICDHLFQHLKRFATEHKSMSVEVIRKAYQATEEGFLGVVTKHWPM 139
VGVYDGHGGPE SRF+ HLFQHL R A ++ ++S E++R+A ATE+GF+ +V + + +
Sbjct: 2 VGVYDGHGGPEASRFVRHHLFQHLMRIAQDYGNISEEILRRAGSATEDGFMKLVHRSYMI 181
Query: 140 NPQIAAVGSCCLVGVICGGSLYIANLGDSRAVLGRAVRATGEVLAIQLSPEHNVAIESVR 199
P IA++ SCC+VGVI G+LYI NLGDSRAV G R + +++ + LSPEH+ E +
Sbjct: 182 KPLIASIASCCMVGVIWKGTLYIGNLGDSRAVAGSICR-SNKIIVVHLSPEHSACWEDIL 358
Query: 200 QEMHSLHPDDPKIVVLKHNVWRV 222
+H + IV + RV
Sbjct: 359 HYSACVHLRNTHIVEMNRGPCRV 427
>BE475454
Length = 424
Score = 130 bits (326), Expect = 1e-30
Identities = 65/148 (43%), Positives = 98/148 (65%), Gaps = 1/148 (0%)
Frame = +2
Query: 56 LLEDQSQIESGPLSFLDTGPYGTFVGVYDGHGGPETSRFICDHLFQHLKRFATE-HKSMS 114
+LEDQSQIESGPL GTFVG+YDGHGGP SR++C+HLF+H + +TE H+ ++
Sbjct: 5 VLEDQSQIESGPL--------GTFVGIYDGHGGPNASRYVCNHLFRHFQAISTESHRVVT 160
Query: 115 VEVIRKAYQATEEGFLGVVTKHWPMNPQIAAVGSCCLVGVICGGSLYIANLGDSRAVLGR 174
+ I +A++ TEEG+ +V+ PQI + + LV VI +L++ N GDS VLG+
Sbjct: 161 PKTIERAFRQTEEGYTSLVSSS*NAQPQIISTKTYYLVKVIFQHTLFMTNTGDSHVVLGK 340
Query: 175 AVRATGEVLAIQLSPEHNVAIESVRQEM 202
+ T ++ I LS EH+ +E+VR+++
Sbjct: 341 KISNTRDITTIHLSAEHSADLETVRRKL 424
>TC221645 similar to UP|Q9LSN8 (Q9LSN8) Protein phosphatase 2C-like protein,
partial (29%)
Length = 680
Score = 124 bits (311), Expect = 7e-29
Identities = 70/162 (43%), Positives = 100/162 (61%), Gaps = 9/162 (5%)
Frame = +1
Query: 1 MLSTLMDFLTACWRRRSSDRKSSDVCGK-----KEGLLWYKDAGQHLFGDYSMAVVQANN 55
ML LM + CW+ +++ K+GLLW++D G+ GD+SMAVVQAN
Sbjct: 217 MLHALMSLIARCWKPFGHGDDAANAAAAAGRECKDGLLWFRDIGKFAAGDFSMAVVQANQ 396
Query: 56 LLEDQSQIESGPLSFLDTGPYGTFVGVYDGHGGPETSRFICDHLFQHLKRFATEHKS-MS 114
+LEDQSQIES G +G+FVGVYDGHGGP+ SR++CD+LF++L+ E +S ++
Sbjct: 397 VLEDQSQIES--------GAFGSFVGVYDGHGGPDCSRYVCDNLFRNLQAILAESQSVVT 552
Query: 115 VEVIRKAYQAT-EEGFLGVVTKHWPMNP--QIAAVGSCCLVG 153
E I++A++A +GF G + P QIA +CCLVG
Sbjct: 553 SEAIQQAFRADWRKGFTGGLFSGAVGAPGRQIATTWTCCLVG 678
>BE804844 similar to GP|13937198|gb| AT4g33920/F17I5_110 {Arabidopsis
thaliana}, partial (23%)
Length = 346
Score = 122 bits (305), Expect = 4e-28
Identities = 61/101 (60%), Positives = 73/101 (71%)
Frame = +2
Query: 266 PSISVHELQEHDQFLIFASDGLWEHLSNQDAVDIVQNHPHSGSARKLIKVALLEAAKKRE 325
PSI + EL+ D FLIFASDGLWE LS++ AV IV HP +G A++L++ AL E A+KRE
Sbjct: 2 PSIIIRELESQDLFLIFASDGLWEQLSDEAAVQIVYKHPRAGIAKRLVRAALHEPAQKRE 181
Query: 326 MRYSDLKKIDRGVRRHFHDDITVVVIFLDSNLVSRASTVTG 366
M Y DL KID G RRHFHDDITVVVI+LD + S T G
Sbjct: 182 MIYDDL*KIDLGFRRHFHDDITVVVIYLDHHARSSKQTAVG 304
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,805,597
Number of Sequences: 63676
Number of extensions: 218635
Number of successful extensions: 1292
Number of sequences better than 10.0: 145
Number of HSP's better than 10.0 without gapping: 1203
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1215
length of query: 392
length of database: 12,639,632
effective HSP length: 99
effective length of query: 293
effective length of database: 6,335,708
effective search space: 1856362444
effective search space used: 1856362444
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC149547.7


