

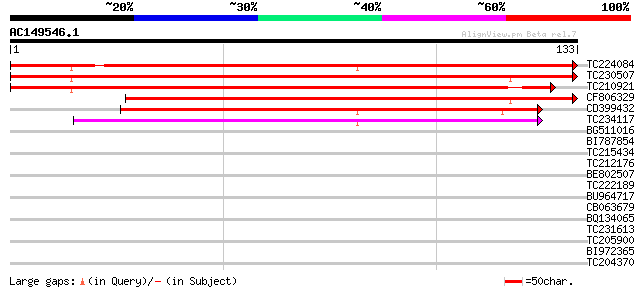
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149546.1 - phase: 0
(133 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC224084 weakly similar to UP|Q93XF3 (Q93XF3) Kinesin heavy chai... 198 5e-52
TC230507 weakly similar to UP|Q9SVJ8 (Q9SVJ8) Kinesin like prote... 131 1e-31
TC210921 weakly similar to UP|Q9SVJ8 (Q9SVJ8) Kinesin like prote... 123 2e-29
CF806329 112 5e-26
CD399432 similar to GP|19570247|db kinesin-like protein NACK1 {N... 82 1e-16
TC234117 similar to GB|CAD42234.1|22796151|ATH495781 kinesin-lik... 70 4e-13
BG511016 29 0.74
BI787854 homologue to SP|O04905|UMPK_ Uridylate kinase (EC 2.7.4... 28 0.96
TC215434 similar to UP|UMPK_ARATH (O04905) Uridylate kinase (UK... 28 0.96
TC212176 similar to UP|Q6ZQ35 (Q6ZQ35) MKIAA0841 protein (Fragme... 28 1.6
BE802507 weakly similar to GP|20197209|gb expressed protein {Ara... 27 2.8
TC222189 similar to UP|ITBX_DROME (P11584) Integrin beta-PS prec... 27 2.8
BU964717 similar to PIR|S61766|S617 protein kinase ATN1 (EC 2.7.... 27 3.7
CB063679 26 4.8
BQ134065 26 4.8
TC231613 similar to UP|ANX4_FRAAN (P51074) Annexin-like protein ... 26 6.2
TC205900 homologue to UP|Q8GSM9 (Q8GSM9) Squalene monooxygenase ... 26 6.2
BI972365 25 8.1
TC204370 weakly similar to GB|AAM19981.1|20466119|AY098971 AT5g4... 25 8.1
>TC224084 weakly similar to UP|Q93XF3 (Q93XF3) Kinesin heavy chain
(Fragment), partial (17%)
Length = 772
Score = 198 bits (504), Expect = 5e-52
Identities = 105/139 (75%), Positives = 119/139 (85%), Gaps = 6/139 (4%)
Frame = +2
Query: 1 MEVELRRLSYLKD-----NQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNM 55
M VELRRL YLK NQ +EDG LTPESS+RYLR ERQMLSRQMQ KLS+SER+++
Sbjct: 188 MGVELRRLFYLKQTFDHWNQTVEDG--LTPESSERYLRGERQMLSRQMQNKLSRSERESL 361
Query: 56 YLKWGISMSSKHRRLQLAHRLWSET-DINHVRESATIVAKLVGTVEPDQAFKEMFGLNFA 114
Y+KWGI +SSK+RRL LAH LWSET D+ H+RESATIVAKLVG+VEPDQAFKEMF LNFA
Sbjct: 362 YIKWGIRLSSKNRRLHLAHCLWSETEDLEHIRESATIVAKLVGSVEPDQAFKEMFVLNFA 541
Query: 115 PRRRRKKSFGWTSSMKHIL 133
PRR RKKSFGWT+SMK+IL
Sbjct: 542 PRRTRKKSFGWTASMKNIL 598
>TC230507 weakly similar to UP|Q9SVJ8 (Q9SVJ8) Kinesin like protein, partial
(17%)
Length = 766
Score = 131 bits (329), Expect = 1e-31
Identities = 73/139 (52%), Positives = 98/139 (69%), Gaps = 6/139 (4%)
Frame = +1
Query: 1 MEVELRRLSYLKD-----NQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNM 55
MEVELRRLS+LK+ N+ + D +T+T SS + LRRER ML + MQR+LS+ ER +
Sbjct: 157 MEVELRRLSFLKESFSDGNKSVRDSQTITLASSVKALRRERGMLVKLMQRRLSEKERRRL 336
Query: 56 YLKWGISMSSKHRRLQLAHRLWSETDINHVRESATIVAKLVGTVEPDQAFKEMFGLNFAP 115
Y + GI++ SK RR+QLA+ LWSE D+NHV +SATIVAKLV E +A KEMFGL+F P
Sbjct: 337 YEECGIALDSKRRRVQLANSLWSENDMNHVMQSATIVAKLVRFWERGKALKEMFGLSFTP 516
Query: 116 R-RRRKKSFGWTSSMKHIL 133
+ R+ S+ W +S +L
Sbjct: 517 QLTGRRSSYPWKNSSASLL 573
>TC210921 weakly similar to UP|Q9SVJ8 (Q9SVJ8) Kinesin like protein, partial
(14%)
Length = 1052
Score = 123 bits (309), Expect = 2e-29
Identities = 65/133 (48%), Positives = 92/133 (68%), Gaps = 5/133 (3%)
Frame = +3
Query: 1 MEVELRRLSYLKD-----NQILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNM 55
MEVELRRLS+LK+ NQ D T+T SS + +R ER++L + M+R+LS+ ER N+
Sbjct: 492 MEVELRRLSFLKETFASGNQSTNDAHTVTLASSAKGVRWEREVLVKLMRRRLSEEERKNL 671
Query: 56 YLKWGISMSSKHRRLQLAHRLWSETDINHVRESATIVAKLVGTVEPDQAFKEMFGLNFAP 115
+ KWGI++ SK RR QLA+R+WS T +NH+ ESA +VAKL+ +A KEMFGL+F+P
Sbjct: 672 FSKWGIALDSKRRRKQLANRIWSSTVMNHIVESAAVVAKLLRFTGQGKALKEMFGLSFSP 851
Query: 116 RRRRKKSFGWTSS 128
R S+ W ++
Sbjct: 852 HR---MSYSWRNT 881
>CF806329
Length = 480
Score = 112 bits (280), Expect = 5e-26
Identities = 58/107 (54%), Positives = 77/107 (71%), Gaps = 1/107 (0%)
Frame = -3
Query: 28 SKRYLRRERQMLSRQMQRKLSKSERDNMYLKWGISMSSKHRRLQLAHRLWSETDINHVRE 87
S + LRRER ML + MQR+LS+ ER +Y +WGI++ SK RR+QL +RLWSE D+NHV +
Sbjct: 478 SVKALRRERGMLVKLMQRRLSEKERRRLYEEWGIALDSKRRRVQLGNRLWSENDMNHVMQ 299
Query: 88 SATIVAKLVGTVEPDQAFKEMFGLNFAPR-RRRKKSFGWTSSMKHIL 133
SATIVAKLV E +A KEMFGL+F P+ R+ S+ W +S +L
Sbjct: 298 SATIVAKLVRFWERGKALKEMFGLSFTPQLTGRRSSYPWKNSSTSLL 158
>CD399432 similar to GP|19570247|db kinesin-like protein NACK1 {Nicotiana
tabacum}, partial (10%)
Length = 390
Score = 81.6 bits (200), Expect = 1e-16
Identities = 44/101 (43%), Positives = 62/101 (60%), Gaps = 2/101 (1%)
Frame = -1
Query: 27 SSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWGISMSSKHRRLQLAHRLWSET-DINHV 85
SS R L+RER+ L++++ +LS ER+ +Y+KW + + K R++Q H+LW++ D HV
Sbjct: 372 SSIRALKREREFLAQRLTSRLSLEEREALYMKWDVPLDGKQRKMQFIHKLWTDPHDQIHV 193
Query: 86 RESATIVAKLVGTVEPDQAFKEMFGLNFA-PRRRRKKSFGW 125
+ESA IVAKLV KEMF LNF P R GW
Sbjct: 192 QESAEIVAKLVSFRTGGNMSKEMFELNFVLPSDNRPCLMGW 70
>TC234117 similar to GB|CAD42234.1|22796151|ATH495781 kinesin-like protein
{Arabidopsis thaliana;} , partial (11%)
Length = 488
Score = 69.7 bits (169), Expect = 4e-13
Identities = 41/111 (36%), Positives = 64/111 (56%), Gaps = 1/111 (0%)
Frame = +1
Query: 16 ILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWGISMSSKHRRLQLAHR 75
IL D + +S R L++ER+ L+R++ L+ ER+ +Y KW + RRLQ ++
Sbjct: 37 ILGDEPAGSVSASIRALKQEREHLARKVNTILTAEERELLYAKWEVPPVGILRRLQFVNK 216
Query: 76 LWSET-DINHVRESATIVAKLVGTVEPDQAFKEMFGLNFAPRRRRKKSFGW 125
LW++ ++ HV+ESA IVAKL+ D K+M LNF+ +K GW
Sbjct: 217 LWTDPYNMQHVQESAEIVAKLIDFSVSDVNSKDMIELNFSSPFNKKTWAGW 369
>BG511016
Length = 431
Score = 28.9 bits (63), Expect = 0.74
Identities = 12/22 (54%), Positives = 16/22 (72%)
Frame = -1
Query: 10 YLKDNQILEDGRTLTPESSKRY 31
YL+DN I++D + TP SSK Y
Sbjct: 266 YLQDNIIIKDTKXTTPASSKAY 201
>BI787854 homologue to SP|O04905|UMPK_ Uridylate kinase (EC 2.7.4.-) (UK)
(Uridine monophosphate kinase) (UMP kinase) (UMP/CMP
kinase)., partial (10%)
Length = 370
Score = 28.5 bits (62), Expect = 0.96
Identities = 13/35 (37%), Positives = 21/35 (59%)
Frame = +1
Query: 21 RTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNM 55
R + + SKRYLR+ +Q L ++M R + D+M
Sbjct: 163 RLMLQDLSKRYLRQSKQFLVQRMTRLIEHVHLDSM 267
>TC215434 similar to UP|UMPK_ARATH (O04905) Uridylate kinase (UK) (Uridine
monophosphate kinase) (UMP kinase) (UMP/CMP kinase) ,
partial (28%)
Length = 540
Score = 28.5 bits (62), Expect = 0.96
Identities = 13/35 (37%), Positives = 21/35 (59%)
Frame = +2
Query: 21 RTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNM 55
R + + SKRYLR+ +Q L ++M R + D+M
Sbjct: 110 RLMLQDLSKRYLRQSKQFLVQRMTRLIEHVHLDSM 214
>TC212176 similar to UP|Q6ZQ35 (Q6ZQ35) MKIAA0841 protein (Fragment), partial
(5%)
Length = 445
Score = 27.7 bits (60), Expect = 1.6
Identities = 16/48 (33%), Positives = 23/48 (47%)
Frame = -2
Query: 22 TLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWGISMSSKHRR 69
TL+ + R+LRR+ LS + K + D W I + KHRR
Sbjct: 198 TLSLKPENRHLRRKVFELSGDVLTKGPDKQLDRFLTDWKIFLCLKHRR 55
>BE802507 weakly similar to GP|20197209|gb expressed protein {Arabidopsis
thaliana}, partial (52%)
Length = 484
Score = 26.9 bits (58), Expect = 2.8
Identities = 11/20 (55%), Positives = 13/20 (65%)
Frame = -1
Query: 111 LNFAPRRRRKKSFGWTSSMK 130
LNF P R R+ GW SS+K
Sbjct: 256 LNFEPLRNRRLPDGWNSSLK 197
>TC222189 similar to UP|ITBX_DROME (P11584) Integrin beta-PS precursor
(Position-specific antigen, beta chain) (Myospheroid
protein) (Olfactory C protein), partial (3%)
Length = 1477
Score = 26.9 bits (58), Expect = 2.8
Identities = 14/56 (25%), Positives = 26/56 (46%)
Frame = +3
Query: 16 ILEDGRTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWGISMSSKHRRLQ 71
+L+DG E + +R Q+ +Q+ ++ E + LKW S K R ++
Sbjct: 1041 VLQDGGKSAWEKKQWMKKRVVQLEEQQVSYQMQAFELEKQRLKWARFSSKKEREME 1208
>BU964717 similar to PIR|S61766|S617 protein kinase ATN1 (EC 2.7.1.-) -
Arabidopsis thaliana, partial (12%)
Length = 378
Score = 26.6 bits (57), Expect = 3.7
Identities = 13/30 (43%), Positives = 16/30 (53%)
Frame = -3
Query: 102 DQAFKEMFGLNFAPRRRRKKSFGWTSSMKH 131
D++F + G RRRRKK F W KH
Sbjct: 115 DESF-DFMG*RRRRRRRRKKMFNWVPPKKH 29
>CB063679
Length = 413
Score = 26.2 bits (56), Expect = 4.8
Identities = 12/26 (46%), Positives = 19/26 (72%)
Frame = +2
Query: 21 RTLTPESSKRYLRRERQMLSRQMQRK 46
+TLT ES+KR RRER ++++R+
Sbjct: 98 KTLTAESNKRKKRRERDDRRQRIERE 175
>BQ134065
Length = 422
Score = 26.2 bits (56), Expect = 4.8
Identities = 12/26 (46%), Positives = 19/26 (72%)
Frame = +2
Query: 21 RTLTPESSKRYLRRERQMLSRQMQRK 46
+TLT ES+KR RRER ++++R+
Sbjct: 173 KTLTAESNKRKKRRERDDRRQRIERE 250
>TC231613 similar to UP|ANX4_FRAAN (P51074) Annexin-like protein RJ4, partial
(64%)
Length = 794
Score = 25.8 bits (55), Expect = 6.2
Identities = 18/45 (40%), Positives = 22/45 (48%), Gaps = 1/45 (2%)
Frame = +1
Query: 17 LEDGRTLTPESSKRYLRRERQM-LSRQMQRKLSKSERDNMYLKWG 60
L G T SKR + RE LS Q RKL ++ R + L WG
Sbjct: 484 LSPGLRRT*RRSKRCITRETVFTLSMQWPRKLLETTRSSSSL*WG 618
>TC205900 homologue to UP|Q8GSM9 (Q8GSM9) Squalene monooxygenase 2 , partial
(36%)
Length = 825
Score = 25.8 bits (55), Expect = 6.2
Identities = 13/35 (37%), Positives = 21/35 (59%)
Frame = -1
Query: 43 MQRKLSKSERDNMYLKWGISMSSKHRRLQLAHRLW 77
M +KL K E MYL W + ++++R+Q R+W
Sbjct: 261 MHKKLCK-EHLPMYLWWMLQAYARYKRIQGICRVW 160
>BI972365
Length = 392
Score = 25.4 bits (54), Expect = 8.1
Identities = 22/70 (31%), Positives = 34/70 (48%), Gaps = 5/70 (7%)
Frame = -1
Query: 41 RQMQRKLSKSERDNMYLKWGISMSSKHRRLQLA-----HRLWSETDINHVRESATIVAKL 95
+Q Q K +KSE+ + L W ++ L+L+ RL +N +R++ IV KL
Sbjct: 317 KQNQNKSTKSEKQKLLLVWCQTIVKCQ*SLELSLDYKLIRLKETKQVNKIRKT-KIVPKL 141
Query: 96 VGTVEPDQAF 105
TV Q F
Sbjct: 140 A*TVAKCQ*F 111
>TC204370 weakly similar to GB|AAM19981.1|20466119|AY098971 AT5g44130/MLN1_5
{Arabidopsis thaliana;} , partial (59%)
Length = 1002
Score = 25.4 bits (54), Expect = 8.1
Identities = 20/85 (23%), Positives = 35/85 (40%)
Frame = -2
Query: 21 RTLTPESSKRYLRRERQMLSRQMQRKLSKSERDNMYLKWGISMSSKHRRLQLAHRLWSET 80
R+ T + LR + L R+ R++S + + HR+ +L + E
Sbjct: 668 RSATSLTHAGNLRSTLRTLRRRSLRRMSGLGGSKELQRQQHLVHLVHRQRKLLPQRVVEL 489
Query: 81 DINHVRESATIVAKLVGTVEPDQAF 105
++H R +V L G V+P F
Sbjct: 488 RLHHARGDVHLVTALAGKVQPPSPF 414
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.131 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,095,842
Number of Sequences: 63676
Number of extensions: 50635
Number of successful extensions: 354
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 348
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 348
length of query: 133
length of database: 12,639,632
effective HSP length: 87
effective length of query: 46
effective length of database: 7,099,820
effective search space: 326591720
effective search space used: 326591720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 53 (25.0 bits)
Medicago: description of AC149546.1


