

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
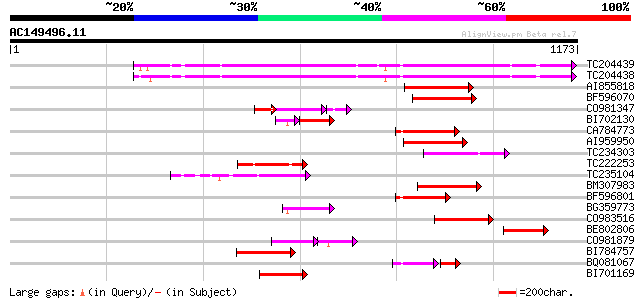
Query= AC149496.11 - phase: 0 /pseudo
(1173 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 453 e-127
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 445 e-125
AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza ... 149 5e-36
BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vi... 138 1e-32
CO981347 91 3e-30
BI702130 weakly similar to GP|4234854|gb| pol polyprotein {Zea m... 102 3e-29
CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {V... 126 5e-29
AI959950 122 1e-27
TC234303 weakly similar to UP|Q8W153 (Q8W153) Polyprotein, parti... 117 2e-26
TC222253 similar to UP|Q9ZQE4 (Q9ZQE4) Copia-like retroelement p... 113 6e-25
TC235104 weakly similar to UP|Q850H8 (Q850H8) Gag-pol polyprotei... 109 8e-24
BM307983 108 1e-23
BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Gl... 105 9e-23
BG359773 104 2e-22
CO983516 104 2e-22
BE802806 103 6e-22
CO981879 76 6e-21
BI784757 99 8e-21
BQ081067 weakly similar to GP|23495377|dbj orf490 {Oryza sativa ... 79 1e-20
BI701169 95 2e-19
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 453 bits (1165), Expect = e-127
Identities = 305/952 (32%), Positives = 475/952 (49%), Gaps = 37/952 (3%)
Frame = +1
Query: 257 KCFYCNNAGHWK*------NYPKYLEDKKNGN-----VPTTSAGIFVIEINMSTSTS--W 303
+C YC GH K +P + N VP A V+ ++ S W
Sbjct: 1507 RCHYCGKYGHIKPFCYHLHGHPHHGTQSSNSRKKMMWVPKHKAVSLVVHTSLRASAKEDW 1686
Query: 304 VLDTGCGSHICTDVQGLRKSRALAKGEVDLRVGNGAKVAALAVGTYVLT-LPSGLLLNLE 362
LD+GC H+ + L + V G+G+K + +G V LPS L
Sbjct: 1687 YLDSGCSRHMTGVKEFLLNIEPCSTSYVTF--GDGSKGKIIGMGKLVHDGLPS-----LN 1845
Query: 363 NCYYVPAISRNIISISCLDKAGFEFTIKNKCCSVYLDN--ILYANANLSNGLYVLDLDMP 420
V ++ N+ISIS L GF C V + +L + + Y+
Sbjct: 1846 KVLLVKGLTANLISISQLCDEGFNVNFTKSECLVTNEKSEVLMKGSRSKDNCYLWTPQET 2025
Query: 421 IYNINAKRLKPNELNPTYLWHCRLGHINENRISKLHKDGF---LDSFDFESYETCRSCLL 477
Y+ K +E+ +WH R GH++ + K+ G + + E C C +
Sbjct: 2026 SYSSTCLSSKEDEVR---IWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQI 2196
Query: 478 GKMTKAPFTG-QGERASDLLGLIHTDVCGPLNITARGGFHYFITLTDDFSRFGYVYLMKH 536
GK K Q + S +L L+H D+ GP+ + + GG Y + DDFSRF +V ++
Sbjct: 2197 GKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIRE 2376
Query: 537 KSESFTFFKEFQNEVENQLGKKIKMLRSDRGGEYLSLEFDNHLKECGILSQLTPPGTPQW 596
KSE+F FKE ++ + IK +RSD G E+ + F GI + + TPQ
Sbjct: 2377 KSETFEVFKELSLRLQREKDCVIKRIRSDHGREFENSRFTEFCTSEGITHEFSAAITPQQ 2556
Query: 597 NGVSERRNRTLLDMVRSMMSHAELPNFLWGYALLTAAYTLNRVP-SKAVEKTPYEIWNGR 655
NG+ ER+NRTL + R M+ ELP LW A+ TA Y NRV + T YEIW GR
Sbjct: 2557 NGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGR 2736
Query: 656 KPNVGHFKIWGCEAYI--KRLMSTKLEQKSEKCFFVGYPKETRGYYFYKPPEGTIVVAKT 713
KP+V HF I+G YI R K++ KS+ F+GY +R Y + T++ +
Sbjct: 2737 KPSVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESIN 2916
Query: 714 GVFLEKDFVSRRISGSKVDLKEIQDPQSTEIPVEEQGQDTQTVMTENPAPVTQEPRRSS- 772
V D R + D++ D + E +++ + E+ + Q +RSS
Sbjct: 2917 VVV--DDLSPARKKDVEEDVRTSGDNVADAAKSGENAENSDSATDESN--INQPDKRSST 3084
Query: 773 RIRQ---------EPERYGYLISQEGDVL----LMDQDEPVTYTEAITGPEFEKWLEAMK 819
RI++ +P R S+E +++ + + EP EA+T E W+ AM+
Sbjct: 3085 RIQKMHPKELIIGDPNRGVTTRSREVEIVSNSCFVSKIEPKNVKEALTD---EFWINAMQ 3255
Query: 820 SEMDSMYTNQVWNLIDAPEGINPIGCKWVFKKKIDMDGKVSTYKARLVAKGFKQIHGVDY 879
E++ N+VW L+ PEG N IG KW+FK K + +G ++ KARLVA+G+ QI GVD+
Sbjct: 3256 EELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDF 3435
Query: 880 DETFSPVAMIKSIRILLAIAAYHDYEIWQMDVKTAFLNGNLLEDVYMTQPEGFGDPKATK 939
DETF+PVA ++SIR+LL +A ++++QMDVK+AFLNG L E+VY+ QP+GF DP
Sbjct: 3436 DETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFADPTHPD 3615
Query: 940 KVCKLQRSIYGLKQASRSWNLRFDETVQQYGFIKNEDEPCVYKKVSGSIVSFLILYVDDI 999
V +L++++YGLKQA R+W R E + Q G+ K + ++ K + +YVDDI
Sbjct: 3616 HVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDI 3795
Query: 1000 LLIGNDIPTLQEIKTWLRKCFSMKDLGEASYILGIRIYRDRSQRLLGLSQGRYIDKVLRR 1059
+ G L+ ++ F M +GE +Y LG+++ + + + LSQ RY ++++
Sbjct: 3796 VFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQV--KQMEDSIFLSQSRYAKNIVKK 3969
Query: 1060 FNMHESKKGFIPMSSGLNLSKTQSPSTDDERDRMRDIPYASAIGSIMYAMICTRPDVSYA 1119
F M + P + L LSK ++ ++ D+ Y S IGS++Y + +RPD++YA
Sbjct: 3970 FGMENASHKRTPAPTHLKLSKDEAGTSVDQS------LYRSMIGSLLY-LTASRPDITYA 4128
Query: 1120 LSATSRYQSNPGNEYWIAVKNILKYLRRTKDTFLIYGG*EELSVIGYIDASF 1171
+ +RYQ+NP + VK ILKY+ T D ++Y ++GY DA +
Sbjct: 4129 VGVCARYQANPKISHLTQVKRILKYVNGTSDYGIMYCHCSNPMLVGYCDADW 4284
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 445 bits (1144), Expect = e-125
Identities = 298/952 (31%), Positives = 472/952 (49%), Gaps = 37/952 (3%)
Frame = +1
Query: 257 KCFYCNNAGHWK*NYPKYLEDKKNGNVPTTSAG--------------IFVIEINMSTSTS 302
+C YC GH K + +L + ++S+G + + S
Sbjct: 1510 RCHYCGKYGHIK-PFCYHLHGHPHHGTQSSSSGRKMMWVPKHKIVSLVVHTSLRASAKED 1686
Query: 303 WVLDTGCGSHICTDVQGLRKSRALAKGEVDLRVGNGAKVAALAVGTYVLT-LPSGLLLNL 361
W LD+GC H+ + L + V G+G+K +G V LPS L
Sbjct: 1687 WYLDSGCSRHMTGVKEFLVNIEPCSTSYVTF--GDGSKGKITGMGKLVHDGLPS-----L 1845
Query: 362 ENCYYVPAISRNIISISCLDKAGFEFTIKNKCCSVYLDN--ILYANANLSNGLYVLDLDM 419
V ++ N+ISIS L GF C V + +L + + Y+
Sbjct: 1846 NKVLLVKGLTANLISISQLCDEGFNVNFTKSECLVTNEKSEVLMKGSRSKDNCYLWTPQE 2025
Query: 420 PIYNINAKRLKPNELNPTYLWHCRLGHINENRISKLHKDGF---LDSFDFESYETCRSCL 476
Y+ K +E+ +WH R GH++ + K+ G + + E C C
Sbjct: 2026 TSYSSTCLFSKEDEVK---IWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQ 2196
Query: 477 LGKMTKAPFTG-QGERASDLLGLIHTDVCGPLNITARGGFHYFITLTDDFSRFGYVYLMK 535
+GK K Q + S +L L+H D+ GP+ + + GG Y + DDFSRF +V ++
Sbjct: 2197 IGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIR 2376
Query: 536 HKSESFTFFKEFQNEVENQLGKKIKMLRSDRGGEYLSLEFDNHLKECGILSQLTPPGTPQ 595
KS++F FKE ++ + IK +RSD G E+ + +F GI + + TPQ
Sbjct: 2377 EKSDTFEVFKELSLRLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEFSAAITPQ 2556
Query: 596 WNGVSERRNRTLLDMVRSMMSHAELPNFLWGYALLTAAYTLNRVP-SKAVEKTPYEIWNG 654
NG+ ER+NRTL + R M+ ELP LW A+ TA Y NRV + T YEIW G
Sbjct: 2557 QNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKG 2736
Query: 655 RKPNVGHFKIWGCEAYI--KRLMSTKLEQKSEKCFFVGYPKETRGYYFYKPPEGTIVVAK 712
RKP V HF I+G YI R K++ KS+ F+GY +R Y + T++ +
Sbjct: 2737 RKPTVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESI 2916
Query: 713 TGVFLEKDFVSRRISGSKVDLKEIQDPQSTEIPVEEQGQDTQTVMTENPAPVTQEPRRSS 772
V D R + D++ D + E +++ + T+ P + R S
Sbjct: 2917 NVVV--DDLTPARKKDVEEDVRTSGDNVADTAKSAENAENSDSA-TDEPNINQPDKRPSI 3087
Query: 773 RIRQ---------EPERYGYLISQEGDVL----LMDQDEPVTYTEAITGPEFEKWLEAMK 819
RI++ +P R S+E +++ + + EP EA+T E W+ AM+
Sbjct: 3088 RIQKMHPKELIIGDPNRGVTTRSREIEIVSNSCFVSKIEPKNVKEALTD---EFWINAMQ 3258
Query: 820 SEMDSMYTNQVWNLIDAPEGINPIGCKWVFKKKIDMDGKVSTYKARLVAKGFKQIHGVDY 879
E++ N+VW L+ PEG N IG KW+FK K + +G ++ KARLVA+G+ QI GVD+
Sbjct: 3259 EELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDF 3438
Query: 880 DETFSPVAMIKSIRILLAIAAYHDYEIWQMDVKTAFLNGNLLEDVYMTQPEGFGDPKATK 939
DETF+PVA ++SIR+LL +A ++++QMDVK+AFLNG L E+ Y+ QP+GF DP
Sbjct: 3439 DETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYVEQPKGFVDPTHPD 3618
Query: 940 KVCKLQRSIYGLKQASRSWNLRFDETVQQYGFIKNEDEPCVYKKVSGSIVSFLILYVDDI 999
V +L++++YGLKQA R+W R E + Q G+ K + ++ K + +YVDDI
Sbjct: 3619 HVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDI 3798
Query: 1000 LLIGNDIPTLQEIKTWLRKCFSMKDLGEASYILGIRIYRDRSQRLLGLSQGRYIDKVLRR 1059
+ G L+ ++ F M +GE +Y LG+++ + + + LSQ +Y ++++
Sbjct: 3799 VFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQV--KQMEDSIFLSQSKYAKNIVKK 3972
Query: 1060 FNMHESKKGFIPMSSGLNLSKTQSPSTDDERDRMRDIPYASAIGSIMYAMICTRPDVSYA 1119
F M + P + L LSK ++ ++ D+ Y S IGS++Y + +RPD++YA
Sbjct: 3973 FGMENASHKRTPAPTHLKLSKDEAGTSVDQS------LYRSMIGSLLY-LTASRPDITYA 4131
Query: 1120 LSATSRYQSNPGNEYWIAVKNILKYLRRTKDTFLIYGG*EELSVIGYIDASF 1171
+ +RYQ+NP + VK ILKY+ T D ++Y + ++GY DA +
Sbjct: 4132 VGVCARYQANPKISHLNQVKRILKYVNGTSDYGIMYCHCSDSMLVGYCDADW 4287
>AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza sativa
(japonica cultivar-group)}, partial (10%)
Length = 463
Score = 149 bits (377), Expect = 5e-36
Identities = 70/142 (49%), Positives = 100/142 (70%)
Frame = -3
Query: 817 AMKSEMDSMYTNQVWNLIDAPEGINPIGCKWVFKKKIDMDGKVSTYKARLVAKGFKQIHG 876
AM+ E++ N VW L++ PE IG KWVF+ K+D G + KARLVAKG+ Q G
Sbjct: 458 AMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIIIRNKARLVAKGYNQEEG 279
Query: 877 VDYDETFSPVAMIKSIRILLAIAAYHDYEIWQMDVKTAFLNGNLLEDVYMTQPEGFGDPK 936
+DY+ET++PVA ++ IR+LLA + +++++QMDVK+AFLNG + E+VY+ QP GF P
Sbjct: 278 IDYEETYAPVARLEVIRMLLAYVSIMNFKLYQMDVKSAFLNGLIQEEVYVEQPPGFEIPD 99
Query: 937 ATKKVCKLQRSIYGLKQASRSW 958
V KLQ+++YGLKQA R+W
Sbjct: 98 KPTHVYKLQKALYGLKQAPRAW 33
>BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vinifera},
partial (34%)
Length = 407
Score = 138 bits (348), Expect = 1e-32
Identities = 64/133 (48%), Positives = 91/133 (68%)
Frame = -2
Query: 834 IDAPEGINPIGCKWVFKKKIDMDGKVSTYKARLVAKGFKQIHGVDYDETFSPVAMIKSIR 893
+ P G P+GC+WV+ K+ G+V KARLVAKG+ Q++G+DY +TFSPVA + ++R
Sbjct: 406 VPLPPGKTPVGCRWVYTVKVGPTGEVDRLKARLVAKGYTQVYGIDYCDTFSPVAKLTTVR 227
Query: 894 ILLAIAAYHDYEIWQMDVKTAFLNGNLLEDVYMTQPEGFGDPKATKKVCKLQRSIYGLKQ 953
+ LA+AA + + Q+D+K AFL+G+L ED+YM QP GF VCKL RS+YGLKQ
Sbjct: 226 LFLAMAAICHWPLHQLDIKNAFLHGDLEEDIYMEQPPGFVAQGEYGLVCKLHRSLYGLKQ 47
Query: 954 ASRSWNLRFDETV 966
+ R+W +F V
Sbjct: 46 SPRAWFGKFSHVV 8
>CO981347
Length = 624
Score = 90.9 bits (224), Expect(3) = 3e-30
Identities = 47/112 (41%), Positives = 67/112 (58%), Gaps = 1/112 (0%)
Frame = +2
Query: 544 FKEFQNEVENQLGKKIKMLRSDRGGEYLSLEFDNHLKECGILSQLTPPGTPQWNGVSERR 603
F+E + NQLG K+K+LR+D G E++ +F+ ++ GI P TP NG++ER
Sbjct: 116 FRE*HTLIGNQLGTKLKVLRTDNGLEFVLEQFNEFCRKIGIKRHKIVPHTP*QNGLAERM 295
Query: 604 NRTLLDMVRSMMSHAELPNFLWGYALLTAAYTLNRVPSKAVE-KTPYEIWNG 654
N T+L+ VR M+ A LP WG A T +Y +NR PS + KTP E W+G
Sbjct: 296 NMTILERVRCMLLSARLPKTFWGEAANTTSYLINRCPSSTLGFKTPMEAWSG 451
Score = 44.3 bits (103), Expect(3) = 3e-30
Identities = 20/46 (43%), Positives = 31/46 (66%)
Frame = +3
Query: 506 PLNITARGGFHYFITLTDDFSRFGYVYLMKHKSESFTFFKEFQNEV 551
P + GG YF+T+ DDFSR ++Y++K+KSES FK +N++
Sbjct: 3 PSRVKTHGGSSYFLTIIDDFSRRVWLYVLKNKSES---FKNSENDI 131
Score = 36.6 bits (83), Expect(3) = 3e-30
Identities = 17/52 (32%), Positives = 28/52 (53%)
Frame = +3
Query: 655 RKPNVGHFKIWGCEAYIKRLMSTKLEQKSEKCFFVGYPKETRGYYFYKPPEG 706
+ PN K++G A+ + KL+ ++ KC F+GYPK + Y +K G
Sbjct: 453 KPPNYSGLKVFGSLAF-DHVKQGKLDARAVKCVFIGYPKGVKRYKLWKLEPG 605
>BI702130 weakly similar to GP|4234854|gb| pol polyprotein {Zea mays},
partial (14%)
Length = 421
Score = 102 bits (253), Expect(2) = 3e-29
Identities = 46/74 (62%), Positives = 60/74 (80%)
Frame = -1
Query: 599 VSERRNRTLLDMVRSMMSHAELPNFLWGYALLTAAYTLNRVPSKAVEKTPYEIWNGRKPN 658
V+ERRNRTLLDMVRSM S+ +LP FLW AL TAAY LNRVP+KAV KTP+E++ G KP+
Sbjct: 250 VAERRNRTLLDMVRSMRSNVKLPQFLWIDALKTAAYILNRVPTKAVSKTPFELFKGWKPS 71
Query: 659 VGHFKIWGCEAYIK 672
+ H ++WGC + ++
Sbjct: 70 LRHIRVWGCPSEVR 29
Score = 46.6 bits (109), Expect(2) = 3e-29
Identities = 26/57 (45%), Positives = 33/57 (57%), Gaps = 9/57 (15%)
Frame = -2
Query: 551 VENQLGKKIKMLRSDRGGEYLSL---------EFDNHLKECGILSQLTPPGTPQWNG 598
VE Q GK+IK++RSDRGGEY F L+E I++Q T PG+P NG
Sbjct: 420 VEKQCGKQIKIVRSDRGGEYYGRYTEDGQAPGSFAKFLQEHEIVAQYTMPGSPDQNG 250
>CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {Vitis
vinifera}, partial (34%)
Length = 409
Score = 126 bits (317), Expect = 5e-29
Identities = 58/131 (44%), Positives = 86/131 (65%)
Frame = +3
Query: 799 PVTYTEAITGPEFEKWLEAMKSEMDSMYTNQVWNLIDAPEGINPIGCKWVFKKKIDMDGK 858
P T EA+ P W +AM EM ++ N W L+ P G +GC+WV+ K+ +GK
Sbjct: 21 PSTIREALDHPG---WRQAMVDEMQALENNGTWELVPLPPGKTTVGCRWVYTVKVGPNGK 191
Query: 859 VSTYKARLVAKGFKQIHGVDYDETFSPVAMIKSIRILLAIAAYHDYEIWQMDVKTAFLNG 918
V KARLVAKG+ Q++G++Y +TFSPV + ++R+ LA+AA + + Q+D+K AFL+G
Sbjct: 192 VDRLKARLVAKGYTQVYGIEYCDTFSPVFFLTTVRLFLAMAAIRHWPLHQLDIKNAFLHG 371
Query: 919 NLLEDVYMTQP 929
+L ED+YM QP
Sbjct: 372 DLEEDIYMEQP 404
>AI959950
Length = 466
Score = 122 bits (305), Expect = 1e-27
Identities = 60/132 (45%), Positives = 88/132 (66%)
Frame = -1
Query: 815 LEAMKSEMDSMYTNQVWNLIDAPEGINPIGCKWVFKKKIDMDGKVSTYKARLVAKGFKQI 874
++AM+ E+D N V L+ P+ +G KW+F K+D DGKV YKARLVAKG+ Q
Sbjct: 397 MKAMQEELDQFQKNNV*KLVKLPKRKKVVGVKWIFCNKLDEDGKVVRYKARLVAKGYSQQ 218
Query: 875 HGVDYDETFSPVAMIKSIRILLAIAAYHDYEIWQMDVKTAFLNGNLLEDVYMTQPEGFGD 934
G+DY +TF+ VA ++ I ILL+ A Y + +++QMDVK+AFLNG + ++VY+ QP GF +
Sbjct: 217 EGIDYPKTFALVARLEVICILLSFATYSNMKLYQMDVKSAFLNGLIQKEVYVEQPPGFEN 38
Query: 935 PKATKKVCKLQR 946
+ V KL +
Sbjct: 37 ETLHQHVFKLNK 2
>TC234303 weakly similar to UP|Q8W153 (Q8W153) Polyprotein, partial (10%)
Length = 558
Score = 117 bits (294), Expect = 2e-26
Identities = 71/179 (39%), Positives = 104/179 (57%), Gaps = 1/179 (0%)
Frame = +1
Query: 857 GKVSTYKARLVAKGFKQIHGVDYDETFSPVAMIKSIRILLAIAAYHDYEIWQMDVKTAFL 916
G + +KARLVAK + Q++G DY TFSPVA + + +L ++A + ++ +D K AFL
Sbjct: 28 GTIDQFKARLVAKSYTQVYGQDYTGTFSPVAKMAYVHLLWSMAVVCHWPLF*LDAKNAFL 207
Query: 917 NGNLLEDVYMTQPEGF-GDPKATKKVCKLQRSIYGLKQASRSWNLRFDETVQQYGFIKNE 975
+G L E+VYM QP GF +++ VC+L RS YGLKQ+ R+W + Y +E
Sbjct: 208 HGYLEEEVYMEQPLGFVAQGESSNMVCQLCRSFYGLKQSPRAWPFLYCGAAIWYD--SHE 381
Query: 976 DEPCVYKKVSGSIVSFLILYVDDILLIGNDIPTLQEIKTWLRKCFSMKDLGEASYILGI 1034
+ V+ S +LI+YVDDI + G+D + +K L F KDLG+ Y LGI
Sbjct: 382 ADHSVFYCHSPQGCIYLIVYVDDIGITGSDQHGIT*LK*XLCCQFQTKDLGKLRYFLGI 558
>TC222253 similar to UP|Q9ZQE4 (Q9ZQE4) Copia-like retroelement pol
polyprotein, partial (4%)
Length = 919
Score = 113 bits (282), Expect = 6e-25
Identities = 66/147 (44%), Positives = 90/147 (60%), Gaps = 3/147 (2%)
Frame = +1
Query: 472 CRSCLLGKMTKAPF-TGQGERASDLLGLIHTDVCGPLNITARGGFHYFITLTDDFSRFGY 530
C +C +GK + F TG+ R LL ++H D+C + I G +YFIT DDFS+ +
Sbjct: 331 CDTCEIGKKHRESFPTGKSWRMKKLLKIVHLDLC-TVEIPTHGDNNYFITFIDDFSKKMW 507
Query: 531 VYLMKHKSESFTFFKEFQNEVENQLGKKIKMLRSDRGGEYLS--LEFDNHLKECGILSQL 588
VY +K KSE+ FK F+ E Q G K+K L D+G EYLS + F+ H GI QL
Sbjct: 508 VYFLKQKSEACNAFKMFKAFAEKQNGCKVKALIIDKGQEYLSYTIFFEKH----GIQHQL 675
Query: 589 TPPGTPQWNGVSERRNRTLLDMVRSMM 615
T TPQ NGV+ER+N+T++DMVR M+
Sbjct: 676 TTKYTPQHNGVTERKNKTIMDMVRCML 756
>TC235104 weakly similar to UP|Q850H8 (Q850H8) Gag-pol polyprotein
(Fragment), partial (28%)
Length = 865
Score = 109 bits (272), Expect = 8e-24
Identities = 85/295 (28%), Positives = 137/295 (45%), Gaps = 6/295 (2%)
Frame = +2
Query: 333 LRVGNGAKVAALAVGTYVLTLPSGLLLNLENCYYVPAISRNIISISCLDKAGFEFTIKNK 392
+ + +G++V A +G T L+L + ++ NI S+S L + +
Sbjct: 20 ITLADGSRVVATGIGHVSPTSS----LSLNSVVFILGCPFNITSLSQLTRF--------R 163
Query: 393 CCSVYLDNILYANANLSNGLYVLDLDMPIYNINAKRLKPN------ELNPTYLWHCRLGH 446
CSV D + G + + I + LKPN + L H RLGH
Sbjct: 164 NCSVTFDANSFVIQECGTGWTI---GVGIESHGLYYLKPNLSWVCSAVTSPKLLHERLGH 334
Query: 447 INENRISKLHKDGFLDSFDFESYETCRSCLLGKMTKAPFTGQGERASDLLGLIHTDVCGP 506
+ +++ + + S + C SC LGK ++ R +IH D+ GP
Sbjct: 335 PHLSKLKIM-----VPSLEKIKDLFCESCQLGKHVRSSXRHVESRVDSPFLVIHXDIWGP 499
Query: 507 LNITARGGFHYFITLTDDFSRFGYVYLMKHKSESFTFFKEFQNEVENQLGKKIKMLRSDR 566
+++ + YF+T D+FS+ V+LMK +SE +F N+++ Q GK IK+LRSD
Sbjct: 500 NRVSSMS-YRYFVTFIDEFSQCTRVFLMKERSEILSFLTSV-NKIKTQFGKTIKILRSDN 673
Query: 567 GGEYLSLEFDNHLKECGILSQLTPPGTPQWNGVSERRNRTLLDMVRSMMSHAELP 621
EY S GIL Q + P TPQ N ++ER+NR L++ R+++ HA P
Sbjct: 674 AKEYFSSVISPFXSAQGILHQFSCPHTPQQNDIAERKNRHLVETARTLLLHANEP 838
>BM307983
Length = 406
Score = 108 bits (271), Expect = 1e-23
Identities = 55/134 (41%), Positives = 85/134 (63%), Gaps = 1/134 (0%)
Frame = +2
Query: 843 IGCKWVFKKKIDMDGKVSTYKARLVAKGFKQIHGVDYDETFSPVAM-IKSIRILLAIAAY 901
+GC+W++ K D + YKARLVAKG+ Q +G+DY+ETF+ I+S A
Sbjct: 2 VGCRWIYTVKY*ADDTLDRYKARLVAKGYIQTYGIDYEETFAQWQK*IQSGSSSP*QQAQ 181
Query: 902 HDYEIWQMDVKTAFLNGNLLEDVYMTQPEGFGDPKATKKVCKLQRSIYGLKQASRSWNLR 961
+E+ Q DVK AFL+G+L E+VYM P G+G KVC+L++++YGLKQ+ R+W R
Sbjct: 182 FGWEMHQFDVKNAFLHGSLEEEVYMEIPPGYGASNGGNKVCRLKKALYGLKQSPRAWFGR 361
Query: 962 FDETVQQYGFIKNE 975
F + + G+ +++
Sbjct: 362 FTQAMLSLGYKQSQ 403
>BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Glycine max},
partial (7%)
Length = 336
Score = 105 bits (263), Expect = 9e-23
Identities = 50/114 (43%), Positives = 75/114 (64%)
Frame = +3
Query: 798 EPVTYTEAITGPEFEKWLEAMKSEMDSMYTNQVWNLIDAPEGINPIGCKWVFKKKIDMDG 857
EP EAI + W+ M+ E++ N VW L++ PE IG KWVF+ K+D G
Sbjct: 3 EPKNIKEAIVD---DNWIIVMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHG 173
Query: 858 KVSTYKARLVAKGFKQIHGVDYDETFSPVAMIKSIRILLAIAAYHDYEIWQMDV 911
+ KARLVAKG+ Q G+DY+ET++PVA +++IR+LLA A+ +++++QMDV
Sbjct: 174 IIIRNKARLVAKGYNQEEGIDYEETYAPVARLEAIRMLLAYASIMNFKLYQMDV 335
>BG359773
Length = 382
Score = 104 bits (260), Expect = 2e-22
Identities = 55/117 (47%), Positives = 70/117 (59%), Gaps = 9/117 (7%)
Frame = -1
Query: 565 DRGGEYLSL---------EFDNHLKECGILSQLTPPGTPQWNGVSERRNRTLLDMVRSMM 615
DRGGEY F +++ GI +Q T G PQ NGVSE+RNR L+DMV SM+
Sbjct: 379 DRGGEYYGRYDETEQYXGPFAKLIQKRGICAQYTMLGIPQ*NGVSEKRNRILMDMVTSML 200
Query: 616 SHAELPNFLWGYALLTAAYTLNRVPSKAVEKTPYEIWNGRKPNVGHFKIWGCEAYIK 672
+ LP LW YAL Y LNRVPSKAV KT +E+W R P++ H +WG + I+
Sbjct: 199 INLTLPISLWMYALKIVMYLLNRVPSKAVPKTHFELWTNRTPSIRHLDVWGFQTEIR 29
>CO983516
Length = 724
Score = 104 bits (260), Expect = 2e-22
Identities = 52/121 (42%), Positives = 79/121 (64%)
Frame = +2
Query: 880 DETFSPVAMIKSIRILLAIAAYHDYEIWQMDVKTAFLNGNLLEDVYMTQPEGFGDPKATK 939
D+ F PVA ++SIR+LL +A ++++QMDVK+AFLNG L E+VY+ QP+GF DP
Sbjct: 356 DKEFHPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFIDPTHPD 535
Query: 940 KVCKLQRSIYGLKQASRSWNLRFDETVQQYGFIKNEDEPCVYKKVSGSIVSFLILYVDDI 999
V +L++++YGLKQA R+W R E + Q G+ K + ++ K + +YVDDI
Sbjct: 536 HVYRLKKALYGLKQAPRAWYERLTELLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDI 715
Query: 1000 L 1000
+
Sbjct: 716 V 718
>BE802806
Length = 285
Score = 103 bits (256), Expect = 6e-22
Identities = 50/94 (53%), Positives = 64/94 (67%)
Frame = -2
Query: 1022 MKDLGEASYILGIRIYRDRSQRLLGLSQGRYIDKVLRRFNMHESKKGFIPMSSGLNLSKT 1081
MKDLG A ILGI I+RDR++ L LSQ Y+ KV+ RF MH+SK P+ LS T
Sbjct: 284 MKDLGSARRILGIDIHRDRAKGELFLSQSNYLKKVVERFRMHQSKPVSTPLGHHTKLSVT 105
Query: 1082 QSPSTDDERDRMRDIPYASAIGSIMYAMICTRPD 1115
Q+P T +ER +M PYA+ +GSIMY M+C+RPD
Sbjct: 104 QAPETAEERSKMNQTPYANGVGSIMYGMVCSRPD 3
>CO981879
Length = 576
Score = 76.3 bits (186), Expect(2) = 6e-21
Identities = 37/97 (38%), Positives = 55/97 (56%)
Frame = -1
Query: 542 TFFKEFQNEVENQLGKKIKMLRSDRGGEYLSLEFDNHLKECGILSQLTPPGTPQWNGVSE 601
+ FK F ++ Q KIK+ RSD G EY + E GI+ Q + TPQ NGV+E
Sbjct: 573 SIFKTFFQMIQTQFQVKIKVFRSDNGREYFNKHLSKXXLENGIIHQSSCVDTPQQNGVAE 394
Query: 602 RRNRTLLDMVRSMMSHAELPNFLWGYALLTAAYTLNR 638
R+NR L ++ R+++ + P + WG A+LT Y N+
Sbjct: 393 RKNRHLXEVARALLFQNKAPKYXWGEAILTGTYLKNK 283
Score = 44.3 bits (103), Expect(2) = 6e-21
Identities = 29/90 (32%), Positives = 42/90 (46%), Gaps = 8/90 (8%)
Frame = -2
Query: 638 RVPSKAVE-KTPYEIWNGRKPN-----VGHFKIWGCEAY--IKRLMSTKLEQKSEKCFFV 689
R+PSK + +TP +++ PN KI+GC + I KLE +++KC FV
Sbjct: 284 RMPSKILNFRTPLDVFTSAFPNNRLSCTLPLKIFGCTVFVHIHEPNQGKLEPRAKKCVFV 105
Query: 690 GYPKETRGYYFYKPPEGTIVVAKTGVFLEK 719
GY +GY + P V F EK
Sbjct: 104 GYAPNQKGYKCFDPTSKKTFVTIDVTFFEK 15
>BI784757
Length = 430
Score = 99.4 bits (246), Expect = 8e-21
Identities = 49/123 (39%), Positives = 77/123 (61%), Gaps = 1/123 (0%)
Frame = +1
Query: 470 ETCRSCLLGKMTKAPFTGQGE-RASDLLGLIHTDVCGPLNITARGGFHYFITLTDDFSRF 528
E C CL K +++ F RA + L +I++DVCGP+ + GG YFI+ D+ +R
Sbjct: 61 EVCDGCLQCKQSRSTFKQNVPIRAKEKLEVIYSDVCGPMQTESLGGNRYFISFIDELTRK 240
Query: 529 GYVYLMKHKSESFTFFKEFQNEVENQLGKKIKMLRSDRGGEYLSLEFDNHLKECGILSQL 588
+VYL++ KS+ F F++F+N + Q G IK+LR++ GGEY+S EF + GI+ ++
Sbjct: 241 VWVYLIRRKSDFFEVFEKFKNMAKKQSGSLIKILRTNGGGEYVSTEFQEFCDQQGIIHEV 420
Query: 589 TPP 591
TPP
Sbjct: 421 TPP 429
>BQ081067 weakly similar to GP|23495377|dbj orf490 {Oryza sativa (japonica
cultivar-group)}, partial (18%)
Length = 430
Score = 78.6 bits (192), Expect(2) = 1e-20
Identities = 40/95 (42%), Positives = 56/95 (58%)
Frame = +1
Query: 792 LLMDQDEPVTYTEAITGPEFEKWLEAMKSEMDSMYTNQVWNLIDAPEGINPIGCKWVFKK 851
LL EP T +A+ P W +AM+++ D++ N+ L P G I CKWVF+
Sbjct: 13 LLFTTAEPSTVKQALISPP---WRQAMQADFDALMENKTLTLTSLPSGKAAIDCKWVFRI 183
Query: 852 KIDMDGKVSTYKARLVAKGFKQIHGVDYDETFSPV 886
K ++ G ++ Y++RLVAKGF G DY ETFSPV
Sbjct: 184 KENLYGTLNRYRSRLVAKGFHLKFGCDYSETFSPV 288
Score = 40.8 bits (94), Expect(2) = 1e-20
Identities = 19/42 (45%), Positives = 29/42 (68%)
Frame = +3
Query: 891 SIRILLAIAAYHDYEIWQMDVKTAFLNGNLLEDVYMTQPEGF 932
+IR++L IA + + + Q+D+ AFL+G L E+VYM Q GF
Sbjct: 300 TIRLILFIALTNHWPLQQVDINNAFLHGLLTEEVYMVQLPGF 425
>BI701169
Length = 407
Score = 94.7 bits (234), Expect = 2e-19
Identities = 50/98 (51%), Positives = 64/98 (65%)
Frame = +2
Query: 518 FITLTDDFSRFGYVYLMKHKSESFTFFKEFQNEVENQLGKKIKMLRSDRGGEYLSLEFDN 577
F DDFSR +VY KHK E F FK+F+ VE + G KIK +RSDRGGE+ S EF
Sbjct: 113 FFLFIDDFSRKTWVYFFKHKLEVFENFKKFKAIVEKESGFKIKAMRSDRGGEF*SNEFQK 292
Query: 578 HLKECGILSQLTPPGTPQWNGVSERRNRTLLDMVRSMM 615
+ + GI L +PQ NGV+ER+NRT+L+M RSM+
Sbjct: 293 YCDDHGIRRPLMVLRSPQQNGVAERKNRTILNMARSML 406
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.137 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 50,010,397
Number of Sequences: 63676
Number of extensions: 682547
Number of successful extensions: 3400
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 3322
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3363
length of query: 1173
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1065
effective length of database: 5,762,624
effective search space: 6137194560
effective search space used: 6137194560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC149496.11


