

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149495.3 + phase: 0
(412 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
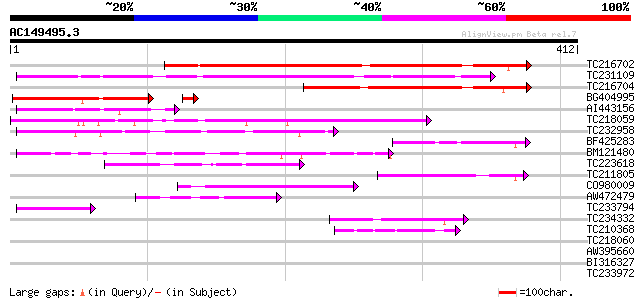
Score E
Sequences producing significant alignments: (bits) Value
TC216702 similar to UP|Q9LDP5 (Q9LDP5) FLORICAULA/LEAFY-like pro... 259 2e-69
TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, par... 251 5e-67
TC216704 similar to UP|Q7PIN1 (Q7PIN1) ENSANGP00000023661 (Fragm... 145 3e-35
BG404995 similar to GP|22831068|dbj OJ1131_E05.32 {Oryza sativa ... 101 1e-22
AI443156 84 2e-16
TC218059 weakly similar to UP|Q84KQ9 (Q84KQ9) F-box, partial (7%) 83 3e-16
TC232958 76 3e-14
BF425283 62 4e-10
BM121480 59 3e-09
TC223618 54 1e-07
TC211805 similar to UP|Q9K6E7 (Q9K6E7) BH3782 protein, partial (... 54 1e-07
CO980009 54 1e-07
AW472479 weakly similar to PIR|T10479|T104 glycine-rich RNA-bind... 51 8e-07
TC233794 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {A... 43 3e-04
TC234332 42 4e-04
TC210368 42 7e-04
TC218060 40 0.001
AW395660 39 0.006
BI316327 37 0.021
TC233972 weakly similar to GB|AAP37713.1|30725382|BT008354 At3g2... 34 0.11
>TC216702 similar to UP|Q9LDP5 (Q9LDP5) FLORICAULA/LEAFY-like protein,
partial (5%)
Length = 1631
Score = 259 bits (662), Expect = 2e-69
Identities = 130/270 (48%), Positives = 184/270 (68%), Gaps = 3/270 (1%)
Frame = +3
Query: 113 NAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKILRLSWL 172
N A++I WNP++R+H I+P+LP+P R P + V GFGFD T DYK++R+S+
Sbjct: 15 NVADDIAFWNPSLRQHRILPYLPVP-RRRHPDTTLFAARVCGFGFDHKTRDYKLVRISYF 191
Query: 173 VSLQNPFYDPHVRLFSLKTNSWKIIPTMPYALVFAQTMGVLVEDSIHWIMAKKLDGLHPS 232
V L + +D V+L++L+ N+WK +P++PYAL A+TMGV V +S+HW++ +KL+ P
Sbjct: 192 VDLHDRSFDAQVKLYTLRANAWKTLPSLPYALCCARTMGVFVGNSLHWVVTRKLEPDQPD 371
Query: 233 LIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLCMTVNYETTKIDVWVMK 292
LI+AF+LT +IF E+PLPD G + EID+A LGG LCMTVN+ T+IDVWVM+
Sbjct: 372 LIIAFDLTHDIFRELPLPDTGGVD-----GGFEIDLALLGGSLCMTVNFHKTRIDVWVMR 536
Query: 293 QYGLKDSWCKLFTMMKSCVTSHLKSSSPLCYSSDGSKVLIEGIEVLLEVHHKKLFWYDLK 352
+Y +DSWCK+FT+ +S LK PL YSSDG+K VLLE K+LFWYDL+
Sbjct: 537 EYNRRDSWCKVFTLEESREMRSLKCVRPLGYSSDGNK-------VLLEHDRKRLFWYDLE 695
Query: 353 TEQVSYEEI---PDFNGAKICVGSLVPPSF 379
++V+ +I P+ N A IC+G+LVPP F
Sbjct: 696 KKEVALVKIQGLPNLNEAMICLGTLVPPYF 785
>TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, partial (8%)
Length = 1061
Score = 251 bits (640), Expect = 5e-67
Identities = 147/348 (42%), Positives = 208/348 (59%)
Frame = +3
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLNRSFILRLRSNIYQ 65
LP + + EI SRLPVKS++R RST K +SIIDS +F+ H S + S ILR RS++Y
Sbjct: 93 LPVEVVTEILSRLPVKSVIRLRSTCKWWRSIIDSRHFVLFHLNKS-HSSLILRHRSHLYS 269
Query: 66 IEDDFSNLTTAVPLNHPFTRNSTNIALIGSCNGLLAVSNGEIALRHPNAANEITIWNPNI 125
+ D S V L+HP S +I ++GS NGLL +SN A++I +WNP +
Sbjct: 270 L-DLKSPEQNPVELSHPLMCYSNSIKVLGSSNGLLCISN---------VADDIALWNPFL 419
Query: 126 RKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKILRLSWLVSLQNPFYDPHVR 185
RKH I+P P + V+GFG + DYK+L +++ V LQ +D V+
Sbjct: 420 RKHRILPADRF----HRPQSSLFAARVYGFGHHSPSNDYKLLSITYFVDLQKRTFDSQVQ 587
Query: 186 LFSLKTNSWKIIPTMPYALVFAQTMGVLVEDSIHWIMAKKLDGLHPSLIVAFNLTLEIFN 245
L++LK++SWK +P+MPYAL A+TMGV V S+HW++ +KL P LIV+F+LT E F+
Sbjct: 588 LYTLKSDSWKNLPSMPYALCCARTMGVFVSGSLHWLVTRKLQPHEPDLIVSFDLTRETFH 767
Query: 246 EVPLPDEIGEEEVNSNDSVEIDVAALGGCLCMTVNYETTKIDVWVMKQYGLKDSWCKLFT 305
EVPLP V N ++ VA LGGCLC+ V + T DVWVM+ YG ++SW KLFT
Sbjct: 768 EVPLP-------VTVNGDFDMQVALLGGCLCV-VEHRGTGFDVWVMRVYGSRNSWEKLFT 923
Query: 306 MMKSCVTSHLKSSSPLCYSSDGSKVLIEGIEVLLEVHHKKLFWYDLKT 353
++++ + S L Y + ++G VL E + KL WY+LKT
Sbjct: 924 LLENNDHHEMMGSGKLKYV---RPLALDGDRVLFEHNRIKLCWYNLKT 1058
>TC216704 similar to UP|Q7PIN1 (Q7PIN1) ENSANGP00000023661 (Fragment),
partial (5%)
Length = 1232
Score = 145 bits (366), Expect = 3e-35
Identities = 81/170 (47%), Positives = 108/170 (62%), Gaps = 4/170 (2%)
Frame = +2
Query: 214 VEDSIHWIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGG 273
V +S+HW++ +KL+ P LIVAF+LT EIF E+PLPD G EIDVA LG
Sbjct: 5 VGNSLHWVVTRKLEPDQPDLIVAFDLTHEIFTELPLPDTGGV-----GGGFEIDVALLGD 169
Query: 274 CLCMTVNYETTKIDVWVMKQYGLKDSWCKLFTMMKSCVTSHLKSS-SPLCYSSDGSKVLI 332
LCMTVN+ +K+DVWVM++Y DSWCKLFT+ +S ++ PL YSSDG+K
Sbjct: 170 SLCMTVNFHNSKMDVWVMREYNRGDSWCKLFTLEESA*VEIVQXCLRPLGYSSDGNK--- 340
Query: 333 EGIEVLLEVHHKKLFWYDLKTEQVS---YEEIPDFNGAKICVGSLVPPSF 379
VLLE K+L WYDL ++V+ + +P+ N A IC+G+LV P F
Sbjct: 341 ----VLLEHDRKRLCWYDLGKKEVTLVRIQGLPNLNEAMICLGTLVTPYF 478
>BG404995 similar to GP|22831068|dbj OJ1131_E05.32 {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 394
Score = 101 bits (251), Expect(2) = 1e-22
Identities = 59/106 (55%), Positives = 70/106 (65%), Gaps = 4/106 (3%)
Frame = +1
Query: 3 SDRLPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSL----NRSFILR 58
SD LP + L EI SRLPV+SLLRFRSTSKS KS+IDS + LH SL N S ILR
Sbjct: 31 SDHLPREVLTEILSRLPVRSLLRFRSTSKSWKSLIDSQHLNWLHLTRSLTLASNTSLILR 210
Query: 59 LRSNIYQIEDDFSNLTTAVPLNHPFTRNSTNIALIGSCNGLLAVSN 104
+ S++YQ +F L V LNHP S +I L+GSCNGLL +SN
Sbjct: 211 VDSDLYQ--TNFPTLDPPVSLNHPLMCYSNSITLLGSCNGLLCISN 342
Score = 23.5 bits (49), Expect(2) = 1e-22
Identities = 7/12 (58%), Positives = 11/12 (91%)
Frame = +3
Query: 126 RKHHIIPFLPLP 137
R+H I+P+LP+P
Sbjct: 345 RQHRILPYLPVP 380
>AI443156
Length = 414
Score = 83.6 bits (205), Expect = 2e-16
Identities = 52/120 (43%), Positives = 70/120 (58%), Gaps = 2/120 (1%)
Frame = +3
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLNRSFILRLRSNIYQ 65
LP + + EI SRLPVKS++R RST K +SIIDS +FI H N + S ILR RS +Y
Sbjct: 90 LPVEVVTEILSRLPVKSVIRLRSTCKWWRSIIDSRHFILFH-LNKSHTSLILRHRSQLYS 266
Query: 66 IEDDFSNLTTAVP--LNHPFTRNSTNIALIGSCNGLLAVSNGEIALRHPNAANEITIWNP 123
+ D +L P L+HP S +I ++GS NGLL +S N ++I +WNP
Sbjct: 267 L--DLKSLLDPNPFELSHPLMCYSNSIKVLGSSNGLLCIS---------NVXDDIALWNP 413
>TC218059 weakly similar to UP|Q84KQ9 (Q84KQ9) F-box, partial (7%)
Length = 1185
Score = 82.8 bits (203), Expect = 3e-16
Identities = 83/332 (25%), Positives = 143/332 (43%), Gaps = 26/332 (7%)
Frame = +3
Query: 1 MASDRLPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRK--NSLN------ 52
M+ + LP + ++ + SRLP K LL + KS +I +F++ + NSL
Sbjct: 51 MSMEHLPGELVSNVLSRLPSKVLLLCKCVCKSWFDLITDPHFVSNYYVVYNSLQSQEEHL 230
Query: 53 ----RSFILRLRSNI----YQIEDDFSNLTTAVPLNHPFTRNSTN---IALIGSCNGLLA 101
R F L++ I + D ++++ V LN P+ NS + ++G CNG+
Sbjct: 231 LVIRRPFFSGLKTYISVLSWNTNDPKKHVSSDV-LNPPYEYNSDHKYWTEILGPCNGIYF 407
Query: 102 VSNGEIALRHPNAANEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLT 161
+ +PN + NP++ + +P SP GFGFDP T
Sbjct: 408 LEG------NPNV-----LMNPSLGEFKALPKSHFT----SPHGTYTFTDYAGFGFDPKT 542
Query: 162 GDYKILRLS--WLVSL-QNPFYDPHVRLFSLKTNSW-KIIPTM---PYALVFAQTMGVLV 214
DYK++ L WL + L+SL +NSW K+ P++ P + + +
Sbjct: 543 NDYKVVVLKDLWLKETDEREIGYWSAELYSLNSNSWRKLDPSLLPLPIEIWGSSRVFTYA 722
Query: 215 EDSIHWIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGC 274
+ HW + +++AF++ E F ++ +P + V + +A G
Sbjct: 723 NNCCHWWGFVEESDATQDVVLAFDMVKESFRKIRVPKIRDSSDEKFGTLVPFEESASIGF 902
Query: 275 LCMTVNYETTKIDVWVMKQYGLKDSWCKLFTM 306
L V + DVWVMK Y + SW K +++
Sbjct: 903 LVYPVRGTEKRFDVWVMKDYWDEGSWVKQYSV 998
>TC232958
Length = 758
Score = 75.9 bits (185), Expect = 3e-14
Identities = 68/251 (27%), Positives = 110/251 (43%), Gaps = 17/251 (6%)
Frame = +1
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLH--------RKNSLNRSFIL 57
LP D + EI RLPVKSL+RF+S KS +I F H + S
Sbjct: 61 LPQDLITEILLRLPVKSLVRFKSVCKSWLFLISDPRFAKSHFDLFAALADRILFIASSAP 240
Query: 58 RLRSNIY--QIEDDFSNLTTAVPLNHPFTRNSTNIALIGSCNGLLAVSNGEIALRHPNAA 115
LRS + + DD +++ V L P + +IGSC G + + +
Sbjct: 241 ELRSIDFNASLHDDSASVAVTVDLPAP-KPYFHFVEIIGSCRGFILL----------HCL 387
Query: 116 NEITIWNPNIRKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKILRLSWLVSL 175
+ + +WNP H ++P P+ D + GFG+DP T D+ ++ +
Sbjct: 388 SHLCVWNPTTGVHKVVPLSPIFFN----KDAVFFTLLCGFGYDPSTDDFLVVHACY---- 543
Query: 176 QNPFYDPH-VRLFSLKTNSWKIIPTMPYALV-FAQT-----MGVLVEDSIHWIMAKKLDG 228
NP + + +FSL+ N+WK I + + F T G + +IHW+ +
Sbjct: 544 -NPKHQANCAEIFSLRANAWKGIEGIHFPYTHFRYTNRYNQFGSFLNGAIHWLAFRINAS 720
Query: 229 LHPSLIVAFNL 239
+ ++IVAF+L
Sbjct: 721 I--NVIVAFDL 747
>BF425283
Length = 336
Score = 62.4 bits (150), Expect = 4e-10
Identities = 41/104 (39%), Positives = 54/104 (51%), Gaps = 4/104 (3%)
Frame = +1
Query: 279 VNYETTKIDVWVMKQYGLKDSWCKLFTMMKSCVTSHLKSSSPLCYSSDGSKVLIEGIEVL 338
V + T VWVM+ YG +DSW KLF++ ++ H S L Y L +G VL
Sbjct: 7 VEHRGTGFHVWVMRVYGSRDSWEKLFSLTEN--HHHEMGSGKLKYVR--PLALDDGDRVL 174
Query: 339 LEVHHKKLFWYDLKTEQVSYEEIPDFNG----AKICVGSLVPPS 378
+ KL WYDLKT VS ++P G +CV SLVPP+
Sbjct: 175 FXHNRXKLCWYDLKTGDVSCVKLPSGIGNTIERTVCVQSLVPPT 306
>BM121480
Length = 917
Score = 59.3 bits (142), Expect = 3e-09
Identities = 75/283 (26%), Positives = 116/283 (40%), Gaps = 9/283 (3%)
Frame = +3
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLNRSFILRLRSNIYQ 65
LP + + EI RLPVKSLLRF+ K+ + L L LRL
Sbjct: 66 LPQELIREILLRLPVKSLLRFKCVL--FKTCSRDVVYFPL----PLPSIPCLRL------ 209
Query: 66 IEDDFSNLTTAVPLNHPFTRNSTNIALIGSCNGLLAVSNGEIALRHPNAANEITIWNPNI 125
DDF ++GSC GL + L + ++AN I +WNP++
Sbjct: 210 --DDFG----------------IRPKILGSCRGL-------VLLYYDDSANLI-LWNPSL 311
Query: 126 RKHHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKILRLSWLVSLQNPFYDPHVR 185
+H +P IT +GFG+D +Y ++ L+ L +
Sbjct: 312 GRHKXLPNYRDDITSFP----------YGFGYDESKDEYLLI----LIGLPKFGPETGAD 449
Query: 186 LFSLKTNSWKI--IPTMPYALVFAQTM----GVLVEDSIHWIMAKKLDGLHPSLIVAFNL 239
++S KT SWK I P A+ G L+ ++HW + + H +I+AF+L
Sbjct: 450 IYSFKTESWKTDTIVYDPLXRYXAEDXIARAGSLLNGALHWFVFSESKXDH--VIIAFDL 623
Query: 240 TLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCL---CMTV 279
+ +PLP + + D+V + +G CL C TV
Sbjct: 624 VERTLSXIPLP--LADRSTVQKDAV-YGLKIMGXCLXVCCSTV 743
>TC223618
Length = 444
Score = 54.3 bits (129), Expect = 1e-07
Identities = 43/147 (29%), Positives = 65/147 (43%), Gaps = 2/147 (1%)
Frame = +1
Query: 70 FSNLTTAVP-LNHPFTRNSTNIALIGSCNGLLAVS-NGEIALRHPNAANEITIWNPNIRK 127
F+NL+T LN+P + ++GSCNGLL + G+ L +WNP+IR
Sbjct: 58 FNNLSTVCDELNYPVKNKFRHDGIVGSCNGLLCFAIKGDCVL----------LWNPSIRV 207
Query: 128 HHIIPFLPLPITPRSPSDMNCSLCVHGFGFDPLTGDYKILRLSWLVSLQNPFYDPHVRLF 187
P L P C G G+D + DYK++ + F + V+++
Sbjct: 208 SKKSPPLGNNWRP------GC-FTAFGLGYDHVNEDYKVVAV--FCDPSEYFIECKVKVY 360
Query: 188 SLKTNSWKIIPTMPYALVFAQTMGVLV 214
S+ TNSW+ I P+ Q G V
Sbjct: 361 SMATNSWRKIQDFPHGFSPFQNSGKFV 441
>TC211805 similar to UP|Q9K6E7 (Q9K6E7) BH3782 protein, partial (13%)
Length = 693
Score = 54.3 bits (129), Expect = 1e-07
Identities = 33/113 (29%), Positives = 54/113 (47%), Gaps = 3/113 (2%)
Frame = +1
Query: 268 VAALGGCLCMTVNYETTKIDVWVMKQYGLKDSWCKLFTMMKSCVTSHLKSSSPLCYSSDG 327
+ L GCLCM +Y+ T VW+MK YG ++SW KL ++ + S P S +G
Sbjct: 4 LGVLQGCLCMNYDYKKTHFVVWMMKDYGARESWVKLVSIPYVPNPENFSYSGPYYISENG 183
Query: 328 SKVLIEGIEVLLEVHHKKLFWYDLKTEQVSYEEIPDFNG---AKICVGSLVPP 377
+L+ +++L Y+ + Y +I G A++ V +LV P
Sbjct: 184 EVLLMFEFDLIL---------YNPRDNSFKYPKIESGKGWFDAEVYVETLVSP 315
>CO980009
Length = 736
Score = 53.9 bits (128), Expect = 1e-07
Identities = 36/134 (26%), Positives = 64/134 (46%), Gaps = 3/134 (2%)
Frame = -2
Query: 123 PNIRKHHIIPFLPLPITPRSPSDMNCSLCVH-GFGFDPLTGDYKILRLSWLVSLQN-PFY 180
P+IR++ +P +P D C + GFD DYK++R+S +V ++
Sbjct: 735 PSIRRYVYLP---------TPHDYPCYHHSYIALGFDSSNCDYKVIRISCIVDDESFGLS 583
Query: 181 DPHVRLFSLKTNSWKIIPTM-PYALVFAQTMGVLVEDSIHWIMAKKLDGLHPSLIVAFNL 239
P L+ L T SW+I+ ++ P V + +HW+ + + G +++F L
Sbjct: 582 APLFELYPLATGSWRILDSISPVCYVAGDAPDGFEDGLVHWVAKRYVTGAWYYFVLSFRL 403
Query: 240 TLEIFNEVPLPDEI 253
E+F EV LP+ +
Sbjct: 402 EDEMFGEVMLPESL 361
>AW472479 weakly similar to PIR|T10479|T104 glycine-rich RNA-binding protein
GRP1 - Commerson's wild potato, partial (22%)
Length = 414
Score = 51.2 bits (121), Expect = 8e-07
Identities = 36/108 (33%), Positives = 57/108 (52%), Gaps = 2/108 (1%)
Frame = +2
Query: 92 LIGSCNGLLAVSNGEIALRHPNAANEITIWNPNIRKHHIIPFLPLPITPRS-PSDMNCSL 150
++ CNGL+ ++ GE L I I NP+IR++ +P TP P N +
Sbjct: 119 VLSFCNGLICIAYGERCL-------PIIICNPSIRRYVWLP------TPHDYPGYYNSCI 259
Query: 151 CVHGFGFDPLTGDYKILRLSWLVSLQN-PFYDPHVRLFSLKTNSWKII 197
+ GFD DYK++R+S +V ++ P V L+SLK+ SW+I+
Sbjct: 260 AL---GFDSTNCDYKVIRISCIVDDESFGLSAPLVELYSLKSGSWRIL 394
>TC233794 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {Arabidopsis
thaliana;} , partial (9%)
Length = 449
Score = 42.7 bits (99), Expect = 3e-04
Identities = 24/57 (42%), Positives = 33/57 (57%)
Frame = +1
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLHRKNSLNRSFILRLRSN 62
LP + + EI RLPVKSLLRF+ KS S+I F+ H + + + L LRS+
Sbjct: 109 LPQELIREILLRLPVKSLLRFKCVCKSFLSLISDPQFVISHYALAASPTHRLILRSH 279
>TC234332
Length = 644
Score = 42.4 bits (98), Expect = 4e-04
Identities = 27/104 (25%), Positives = 49/104 (46%), Gaps = 3/104 (2%)
Frame = +2
Query: 233 LIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLCMTVNYETTKIDVWVMK 292
+I +++L E + + +PD + E + + V L GCLC++ + W+MK
Sbjct: 110 VIFSYDLKNESYRYLLMPDGLLEVPHSPPELV-----VLKGCLCLSHRHGGNHFGFWLMK 274
Query: 293 QYGLKDSWCKLFTMMKSCVTSH---LKSSSPLCYSSDGSKVLIE 333
++G++ SW + + + H L LC S D VL+E
Sbjct: 275 EFGVEKSWTRFLNISYDQLHIHGGFLDHPVILCMSEDDGVVLLE 406
>TC210368
Length = 815
Score = 41.6 bits (96), Expect = 7e-04
Identities = 26/91 (28%), Positives = 43/91 (46%)
Frame = +1
Query: 237 FNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLCMTVNYETTKIDVWVMKQYGL 296
F+L +E+PLP + + D+V + +GGCL + + ++WVMK+Y +
Sbjct: 1 FDLVERTLSEIPLP--LADRSTVQKDAV-YGLRIMGGCLSVCCS-SCGMTEIWVMKEYKV 168
Query: 297 KDSWCKLFTMMKSCVTSHLKSSSPLCYSSDG 327
+ SW F + S SP+C DG
Sbjct: 169 RSSWTMTFLIHTS------NRISPICTIKDG 243
>TC218060
Length = 901
Score = 40.4 bits (93), Expect = 0.001
Identities = 23/88 (26%), Positives = 39/88 (44%)
Frame = +2
Query: 219 HWIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPDEIGEEEVNSNDSVEIDVAALGGCLCMT 278
HW + G +++AF++ E F ++ +P + V + +A G L
Sbjct: 29 HWWGFVEDXGATQDIVLAFDMVKESFRKIRVPKVRDSSDEKFATLVPFEESASIGVLVYP 208
Query: 279 VNYETTKIDVWVMKQYGLKDSWCKLFTM 306
V DVWVMK Y + SW K +++
Sbjct: 209 VRGAEKSFDVWVMKDYWDEGSWVKQYSV 292
>AW395660
Length = 381
Score = 38.5 bits (88), Expect = 0.006
Identities = 19/32 (59%), Positives = 23/32 (71%)
Frame = +3
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSII 37
LP + + EI SRLPVKSLL+FR KS S+I
Sbjct: 264 LPDELVVEILSRLPVKSLLQFRCVCKSWMSLI 359
>BI316327
Length = 410
Score = 36.6 bits (83), Expect = 0.021
Identities = 23/91 (25%), Positives = 43/91 (46%), Gaps = 5/91 (5%)
Frame = +1
Query: 159 PLTGDYKILRLSWLVSLQNPFYDPHVRLFSLKTNSWKIIPTMPYALVFA-----QTMGVL 213
P T DY ++ S+ + H FSL+ N+WK ++ +G
Sbjct: 91 PTTDDYLVVLASYNRNFPQDELVTHFEYFSLRANTWKATDGTGFSYKQCYYYNDNQIGCF 270
Query: 214 VEDSIHWIMAKKLDGLHPSLIVAFNLTLEIF 244
+++HW+ + + L ++IVAF+LT ++F
Sbjct: 271 SNNALHWLAFRFDESL--NVIVAFDLTKKVF 357
>TC233972 weakly similar to GB|AAP37713.1|30725382|BT008354 At3g23880
{Arabidopsis thaliana;} , partial (10%)
Length = 631
Score = 34.3 bits (77), Expect = 0.11
Identities = 18/41 (43%), Positives = 24/41 (57%)
Frame = -2
Query: 6 LPPDTLAEIFSRLPVKSLLRFRSTSKSLKSIIDSHNFINLH 46
LP + + EI RLPVKSL F+ KS S+I + +F H
Sbjct: 540 LPQEIIIEILLRLPVKSLPSFKFVCKS*LSLISNPHFAKWH 418
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,159,033
Number of Sequences: 63676
Number of extensions: 356647
Number of successful extensions: 1909
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 1873
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1884
length of query: 412
length of database: 12,639,632
effective HSP length: 100
effective length of query: 312
effective length of database: 6,272,032
effective search space: 1956873984
effective search space used: 1956873984
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC149495.3


