

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149494.9 + phase: 0
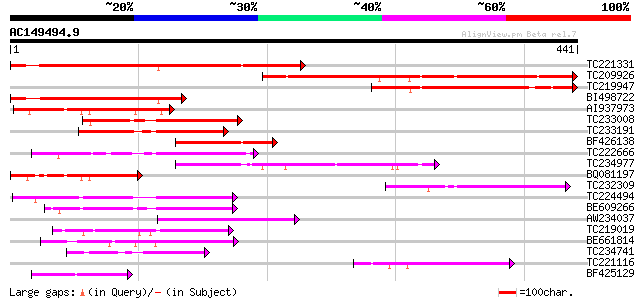
(441 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC221331 homologue to UP|Q9FIT1 (Q9FIT1) Emb|CAB87280.1 (AT5g620... 290 7e-79
TC209926 similar to UP|Q9FIT1 (Q9FIT1) Emb|CAB87280.1 (AT5g62070... 274 5e-74
TC219947 188 4e-48
BI498722 homologue to GP|15215590|gb| AT5g62070/mtg10_90 {Arabid... 159 3e-39
AI937973 homologue to GP|15215590|gb| AT5g62070/mtg10_90 {Arabid... 119 2e-27
TC233008 similar to UP|Q9ASW3 (Q9ASW3) AT3g49260/F2K15_120 (Guar... 117 1e-26
TC233191 similar to UP|Q9LK76 (Q9LK76) Arabidopsis thaliana geno... 111 7e-25
BF426138 110 1e-24
TC222666 similar to UP|Q9MAM4 (Q9MAM4) T25K16.10, partial (22%) 104 9e-23
TC234977 101 8e-22
BQ081197 100 2e-21
TC232309 90 2e-18
TC224494 85 7e-17
BE609266 84 1e-16
AW234037 83 2e-16
TC219019 69 4e-12
BE661814 similar to PIR|A86332|A863 F6F9.8 protein - Arabidopsis... 66 4e-11
TC234741 weakly similar to UP|Q93ZH7 (Q93ZH7) AT5g03040/F15A17_7... 64 1e-10
TC221116 similar to UP|Q9LK76 (Q9LK76) Arabidopsis thaliana geno... 64 2e-10
BF425129 62 4e-10
>TC221331 homologue to UP|Q9FIT1 (Q9FIT1) Emb|CAB87280.1
(AT5g62070/mtg10_90), partial (24%)
Length = 752
Score = 290 bits (743), Expect = 7e-79
Identities = 164/233 (70%), Positives = 181/233 (77%), Gaps = 3/233 (1%)
Frame = +1
Query: 1 MGFLRRLFGAKKPIPPSDGSGKKSDKDNKRRWSFGKQSSKTKSLPQPPP-SAFNQFDSST 59
MGFLRR+FGAKKP K K+RW+F K + + KSLP PPP SA FDSST
Sbjct: 82 MGFLRRIFGAKKP---------SDSKSAKKRWTFLKHTVRNKSLPPPPPPSAVTYFDSST 234
Query: 60 PLERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPE--ETA 117
PL+ NKHAIAVAAATAAVAEAALAAAHAAAEVVRLTS GV+ +S + ETA
Sbjct: 235 PLDANKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSGGVSATSTRPAAMAARVGNLETA 414
Query: 118 AVKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASR 177
AV+IQSAFRGYLARRALRALKALVKLQALVRGHIVRK++ADMLRRMQTLVRLQ +ARASR
Sbjct: 415 AVRIQSAFRGYLARRALRALKALVKLQALVRGHIVRKQSADMLRRMQTLVRLQAQARASR 594
Query: 178 AHLSSDNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKI 230
AHL SD +F SSLSHYPVPEEYE P +STKF GS ILKRCSSN+N R +
Sbjct: 595 AHL-SDPSFNFNSSLSHYPVPEEYEHPPRGFSTKFDGSFILKRCSSNANSRNV 750
>TC209926 similar to UP|Q9FIT1 (Q9FIT1) Emb|CAB87280.1 (AT5g62070/mtg10_90),
partial (9%)
Length = 1271
Score = 274 bits (701), Expect = 5e-74
Identities = 158/274 (57%), Positives = 183/274 (66%), Gaps = 29/274 (10%)
Frame = +3
Query: 197 VPEEYEQPHHVYSTKFGGSSILKRCSSNSNFRKIESEKPRFGSNWLDHWMQENSISQTKN 256
VPE+Y+ YSTKF GS ILKRC SN+NFR I+ EK RFGS+WLD WM+ENS QT++
Sbjct: 252 VPEDYQHSLRAYSTKFDGS-ILKRCRSNANFRDIDVEKARFGSHWLDSWMEENSWRQTRD 428
Query: 257 ASSKNRHPDEHKSDKILEVDTWKPQLNKNE------------------NNVNSMSNESPS 298
AS KN H D+ KSDKILEVDTWKP LN + NN N ++ ESPS
Sbjct: 429 ASLKNGHLDDEKSDKILEVDTWKPHLNSHHSSGSSFQTSSHHYLSSDYNNENFVAYESPS 608
Query: 299 KHSTKAQNQSL---------SVKFHKAKEEVAASRTADNSPQTFSASSRNGSGVRRNTPF 349
K S+KA N SL S+K HK KEE AA + ++SPQ +ASSR GS RR PF
Sbjct: 609 KRSSKALNPSLSSREVLPFGSLKPHKGKEE-AALQNVEDSPQA*TASSRLGSDARRG-PF 782
Query: 350 TPTRSECSWSFLGGYSGYPNYMANTESSRAKVRSQSAPRQRHEFEEYS-STRRPFQGLWD 408
TPT+SEC+WSF GY G+PNYMANTESSRAKVRS SAPRQR EFE Y STRR QGLW+
Sbjct: 783 TPTKSECAWSFFSGYPGHPNYMANTESSRAKVRSHSAPRQRMEFERYGHSTRRSLQGLWE 962
Query: 409 VGSTNSDNDSDSRSNKVSPALSR-FNRIGSSNLR 441
G +SD DSD RS + S NRIGS+NLR
Sbjct: 963 AG-PSSDRDSDFRSKAYATTTSSCLNRIGSANLR 1061
>TC219947
Length = 772
Score = 188 bits (478), Expect = 4e-48
Identities = 109/171 (63%), Positives = 125/171 (72%), Gaps = 11/171 (6%)
Frame = +1
Query: 282 LNKNENNVN-SMSNESPSKHSTKAQNQSLS---------VKFHKAKEEVAASRTADNSPQ 331
LN + NN N ++++ESPSK S KA NQS S +KFH+ KEE A+SRTADNSPQ
Sbjct: 37 LNADYNNENFAVAHESPSKRSAKALNQSFSSREVLQLSSLKFHQGKEE-ASSRTADNSPQ 213
Query: 332 TFSASSRNGSGVRRNT-PFTPTRSECSWSFLGGYSGYPNYMANTESSRAKVRSQSAPRQR 390
FSA SRNGSG RR PFTPTRSECS F GY+G+PNYMANTES RAKVRSQSAPRQR
Sbjct: 214 AFSAHSRNGSGARRGGGPFTPTRSECSLGFFSGYTGHPNYMANTESFRAKVRSQSAPRQR 393
Query: 391 HEFEEYSSTRRPFQGLWDVGSTNSDNDSDSRSNKVSPALSRFNRIGSSNLR 441
EF+ Y STRRP + +NSD+DSD R NKV PA ++ NRIGSSNLR
Sbjct: 394 LEFDRYGSTRRPVH----LVGSNSDHDSDLR-NKVLPAFNQMNRIGSSNLR 531
>BI498722 homologue to GP|15215590|gb| AT5g62070/mtg10_90 {Arabidopsis
thaliana}, partial (14%)
Length = 421
Score = 159 bits (401), Expect = 3e-39
Identities = 97/144 (67%), Positives = 106/144 (73%), Gaps = 7/144 (4%)
Frame = +1
Query: 1 MGFLRRLFGAKKPIPPSDGSGKKSD-KDNKRRWSFGKQSSKTKSLPQPP-PSAFN-QFDS 57
MGFLRRLFGAKKP SD K K+RW+F K + + KSLP PP P A N FDS
Sbjct: 19 MGFLRRLFGAKKP----------SDCKSGKKRWTFLKHTVRNKSLPPPPIPPALNTHFDS 168
Query: 58 STPLERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPE--- 114
STPL+ NKHAIAVAAATAAVAEAALAAAHAAAEVVRLTS GVA +S +R +
Sbjct: 169 STPLDANKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSGGVAATSTSSRTAVTAARVGN 348
Query: 115 -ETAAVKIQSAFRGYLARRALRAL 137
ETAAV+IQSAFRGYLARRALRAL
Sbjct: 349 LETAAVRIQSAFRGYLARRALRAL 420
>AI937973 homologue to GP|15215590|gb| AT5g62070/mtg10_90 {Arabidopsis
thaliana}, partial (12%)
Length = 425
Score = 119 bits (299), Expect = 2e-27
Identities = 82/142 (57%), Positives = 90/142 (62%), Gaps = 17/142 (11%)
Frame = +3
Query: 4 LRRLFGAKKPI---PPSDGSGKKSDKDNKRRWSFGKQSSKTKSLPQPPPSAFNQ---FDS 57
LRRLFG KK PPS K +KDNK+ WSF K S++ K P P+ N FDS
Sbjct: 6 LRRLFGGKKHHNNPPPSSSDASKPNKDNKKTWSFIKHSTRYK--PNTLPTTLNNNSNFDS 179
Query: 58 STP-------LERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTS--SGVAGSSNKTRG 108
ST L+ NKHAIAVAAATAAVAEAALAAAHAAAEVVRLTS GV+
Sbjct: 180 STSSAPFTESLDANKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSGTGGVSSRPAPAPQ 359
Query: 109 QLRLPEET--AAVKIQSAFRGY 128
R+ EET AAVKIQSAFRGY
Sbjct: 360 PRRVAEETTAAAVKIQSAFRGY 425
>TC233008 similar to UP|Q9ASW3 (Q9ASW3) AT3g49260/F2K15_120 (Guard cell
associated protein) (SF16-like protein), partial (29%)
Length = 700
Score = 117 bits (292), Expect = 1e-26
Identities = 75/127 (59%), Positives = 86/127 (67%), Gaps = 2/127 (1%)
Frame = +1
Query: 57 SSTPL--ERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPE 114
+STP+ E HAIAVAAATAA AE A+ AA AAA VVRL +G G +K E
Sbjct: 307 TSTPVTNEDRSHAIAVAAATAAAAETAVVAAQAAARVVRL--AGSYGRQSK--------E 456
Query: 115 ETAAVKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKAR 174
E AA IQS +RGYLARRALRALK LV+LQALVRGH VRK+ +R MQ LVR+Q + R
Sbjct: 457 ERAATLIQSYYRGYLARRALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQARVR 636
Query: 175 ASRAHLS 181
A R LS
Sbjct: 637 ARRFQLS 657
>TC233191 similar to UP|Q9LK76 (Q9LK76) Arabidopsis thaliana genomic DNA,
chromosome 3, P1 clone: MDC8, partial (23%)
Length = 411
Score = 111 bits (277), Expect = 7e-25
Identities = 67/117 (57%), Positives = 82/117 (69%)
Frame = +3
Query: 54 QFDSSTPLERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLP 113
Q D+ E+ KHA+AVAAATA A+AA+AAA A A V+RLTS+ SN T +
Sbjct: 84 QTDTDIQNEQRKHAMAVAAATAVAADAAVAAAQAVAAVIRLTST-----SNATSKSI--- 239
Query: 114 EETAAVKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQ 170
EE AA+KIQSAFR +LA++AL AL+ LVKLQALVRGH+VRK+ LR MQ LV Q
Sbjct: 240 EEAAAIKIQSAFRSHLAKKALCALRGLVKLQALVRGHLVRKQAKATLRCMQALVTAQ 410
>BF426138
Length = 438
Score = 110 bits (275), Expect = 1e-24
Identities = 60/79 (75%), Positives = 67/79 (83%)
Frame = +2
Query: 130 ARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLSSDNLHSFK 189
ARRALRALKALVKLQALVRGHIVRK++ADMLRRMQTLVRLQ +ARASRAHL SD +F
Sbjct: 74 ARRALRALKALVKLQALVRGHIVRKQSADMLRRMQTLVRLQAQARASRAHL-SDPSFNFN 250
Query: 190 SSLSHYPVPEEYEQPHHVY 208
SSLSHYPV + + HH +
Sbjct: 251 SSLSHYPVCKVFYNCHHSF 307
>TC222666 similar to UP|Q9MAM4 (Q9MAM4) T25K16.10, partial (22%)
Length = 1019
Score = 104 bits (259), Expect = 9e-23
Identities = 85/186 (45%), Positives = 99/186 (52%), Gaps = 10/186 (5%)
Frame = +2
Query: 18 DGSGKKSDKDNKRRWSFGK----------QSSKTKSLPQPPPSAFNQFDSSTPLERNKHA 67
D + K KRRW F K S+ T A + S T ++ KHA
Sbjct: 5 DQEEDEEKKREKRRWIFRKTHMSHEGVNNNSNHTTQQKVQHDVAASGGGSRTDQDQ-KHA 181
Query: 68 IAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSNKTRGQLRLPEETAAVKIQSAFRG 127
+AVA ATA EAA+A A AAAEV RL+ K R E AAV IQ+AFRG
Sbjct: 182 LAVAMATA---EAAMATAQAAAEVARLS---------KPASHAR--EHFAAVVIQTAFRG 319
Query: 128 YLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLSSDNLHS 187
YLARRALRALK LVKLQALVRGH VRK+ LR MQ LVR+Q + R S + S
Sbjct: 320 YLARRALRALKGLVKLQALVRGHNVRKQAKMTLRCMQALVRVQARVLDQRIRSSLEG--S 493
Query: 188 FKSSLS 193
KS+ S
Sbjct: 494 RKSTFS 511
>TC234977
Length = 808
Score = 101 bits (251), Expect = 8e-22
Identities = 80/219 (36%), Positives = 120/219 (54%), Gaps = 14/219 (6%)
Frame = +2
Query: 130 ARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRAHLSSDNLHSFK 189
AR+ALRALK LVKLQALVRGHI RK+TA+ L+R+Q L+R+Q + RA RA + LHS
Sbjct: 176 ARKALRALKGLVKLQALVRGHIERKRTAEWLQRVQALLRVQAQIRAGRAQI----LHS-P 340
Query: 190 SSLSHY---PVPEEYEQPHHVYSTKFG--GSSILKRCSSNSNFRKIESEKPRFGSNWLDH 244
SS SH P+++E P S K+ S +LKR SS S + + R S D
Sbjct: 341 SSTSHLRGPATPDKFEIPIRSESMKYDQYSSPLLKRNSSKSRVQINGGNQERCRSRSSDS 520
Query: 245 WMQENSISQTKNASSKNRHPDEHKSDKILEVDTWKPQLNKNENNVNSMSNES--PSKH-- 300
+ E +Q + + ++ DE +S +ILE+D+ KP + N+ +++ S H
Sbjct: 521 RIDEQPWTQ-RRSWTRGCSMDEERSVRILEIDSVKPHVTSKRRNLFYSPSQAMVVSDHYS 697
Query: 301 -----STKAQNQSLSVKFHKAKEEVAASRTADNSPQTFS 334
+T + + +K ++ +E ++ ADNSPQ S
Sbjct: 698 GCNLTTTSPSSYNSPLKINELEE--SSFCAADNSPQALS 808
>BQ081197
Length = 421
Score = 99.8 bits (247), Expect = 2e-21
Identities = 70/116 (60%), Positives = 75/116 (64%), Gaps = 13/116 (11%)
Frame = +2
Query: 1 MGFLRRLFGAKK---PIPPSDGSGKKSDKDNKRRWSFGKQSSKTKSLPQPPPSAFNQ--- 54
MGFLRRLFG KK P P SD S K KD K+ WSF K S++ K P P+ N
Sbjct: 59 MGFLRRLFGGKKHHNPPPSSDAS--KPSKD-KKTWSFVKHSTRYK--PNTLPTTLNNNSN 223
Query: 55 FDSSTP-------LERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSS 103
FD ST L+ NKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSS SS
Sbjct: 224 FDPSTSSSPFPESLDANKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSTGCASS 391
>TC232309
Length = 797
Score = 90.1 bits (222), Expect = 2e-18
Identities = 62/147 (42%), Positives = 82/147 (55%), Gaps = 3/147 (2%)
Frame = +2
Query: 293 SNESPSKHSTKAQNQSLSVKFHKAKEEVAASR--TADNSPQTFSASSRNGSGVRRNTPFT 350
S S S + QS S K EV S TADNSPQ SASS++ G +R+ PFT
Sbjct: 74 STSLQSAQSPCCEVQSHSYSSQKVNNEVEESPFCTADNSPQYLSASSKD-DGFKRS-PFT 247
Query: 351 PTRSECSWSFLGGYSGYPNYMANTESSRAKVRSQSAPRQRHEFEEYSSTRRPFQGLWDVG 410
PTRS+ S S++ GY YP+YMA TESS+AK RS SAP+QR + E S+ R +D+
Sbjct: 248 PTRSDGSRSYIRGYPDYPSYMACTESSKAKARSLSAPKQRPQSERSGSSDRYSLNGFDMS 427
Query: 411 STNSDNDSD-SRSNKVSPALSRFNRIG 436
+ S +NK P R +++G
Sbjct: 428 RLATQRAMQASFTNKAYPGSGRLDKLG 508
>TC224494
Length = 568
Score = 84.7 bits (208), Expect = 7e-17
Identities = 69/178 (38%), Positives = 87/178 (48%), Gaps = 3/178 (1%)
Frame = +2
Query: 3 FLRRLFGAKKPIPPSD--GSGKKSDKDNKRRWSFGKQSSKTKSLPQPPPSAFNQFDSSTP 60
+++ L G KKP G KS K R S G K SA S
Sbjct: 89 WVKALIGLKKPDKEEHVKEGGGKSKKWRLWRSSSGDTGVSWKGFKGGNHSAVAS--SEVG 262
Query: 61 LERNKHAIAVAAATAAVAEAALAAA-HAAAEVVRLTSSGVAGSSNKTRGQLRLPEETAAV 119
+ + H +A AAAT A AA+A A + RL +E AA+
Sbjct: 263 SDSSPHVVAAAAATGAAFTAAVATVVRAPPKDFRLVK-----------------QEWAAI 391
Query: 120 KIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASR 177
+IQ+AFR LARRALRALK +V++QALVRG VRK+ A LR MQ LVR+Q + RA R
Sbjct: 392 RIQTAFRALLARRALRALKGVVRIQALVRGRQVRKQAAVTLRCMQALVRVQARVRARR 565
>BE609266
Length = 446
Score = 84.3 bits (207), Expect = 1e-16
Identities = 60/154 (38%), Positives = 83/154 (52%), Gaps = 4/154 (2%)
Frame = +2
Query: 28 NKRRWSFGKQ----SSKTKSLPQPPPSAFNQFDSSTPLERNKHAIAVAAATAAVAEAALA 83
+K++W F KQ S T PP + T +E + V ATA AE +
Sbjct: 2 SKKKW-FQKQKLQTSESTSQSDNAPPLPLPEI-ILTHVESEINHDRVEVATAVDAEEPVL 175
Query: 84 AAHAAAEVVRLTSSGVAGSSNKTRGQLRLPEETAAVKIQSAFRGYLARRALRALKALVKL 143
A AA V+ T+ + NK EE AA++IQ AFRGYLARRALRAL+ LV+L
Sbjct: 176 AVQTAAAEVQATT--IVQFDNKPT------EEMAAIRIQKAFRGYLARRALRALRGLVRL 331
Query: 144 QALVRGHIVRKKTADMLRRMQTLVRLQTKARASR 177
++L+ G +V+++ LR MQT LQT+ R+ R
Sbjct: 332 RSLMEGPVVKRQAISTLRSMQTFAHLQTQIRSRR 433
>AW234037
Length = 410
Score = 83.2 bits (204), Expect = 2e-16
Identities = 50/110 (45%), Positives = 64/110 (57%)
Frame = +1
Query: 116 TAAVKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARA 175
+A + ++R ARRALRALK LVKLQALVRGHI RK+TA+ L+R+Q L+ QT+ A
Sbjct: 37 SAQCSLSFSYRNC*ARRALRALKGLVKLQALVRGHIERKRTAEWLKRLQALLHAQTQVSA 216
Query: 176 SRAHLSSDNLHSFKSSLSHYPVPEEYEQPHHVYSTKFGGSSILKRCSSNS 225
+S + S L PE++E P S K S ILKR S S
Sbjct: 217 GLTLHASPSSSKLSSHLQGPETPEKFESPIRSKSMKHEHSPILKRNGSKS 366
>TC219019
Length = 796
Score = 68.9 bits (167), Expect = 4e-12
Identities = 57/151 (37%), Positives = 82/151 (53%), Gaps = 10/151 (6%)
Frame = +2
Query: 34 FGKQSSKTKSLPQPPPSAFNQFD----SSTPLERNKHAIAVAAATAAVAEAALAAAHAAA 89
FGK+SSK+ ++ + NQ + +S LE A+ + T A E L + A
Sbjct: 155 FGKKSSKS-NISKGREKLVNQEEGVVVTSKVLETGL-ALEPTSDTIARHEEDLELENEEA 328
Query: 90 EVVRLTSSGV--AGSSNKTRG----QLRLPEETAAVKIQSAFRGYLARRALRALKALVKL 143
E V + + GS N+ ++RL E AA K Q+AFRGYLARRA RALK +++L
Sbjct: 329 ENVIPGNQEIDTVGSINEDAALDPEKIRLEE--AATKAQAAFRGYLARRAFRALKGIIRL 502
Query: 144 QALVRGHIVRKKTADMLRRMQTLVRLQTKAR 174
QAL+RGH+VR++ L M +V+ Q R
Sbjct: 503 QALIRGHLVRRQAVATLCSMYGIVKFQALVR 595
>BE661814 similar to PIR|A86332|A863 F6F9.8 protein - Arabidopsis thaliana,
partial (9%)
Length = 626
Score = 65.9 bits (159), Expect = 4e-11
Identities = 52/165 (31%), Positives = 81/165 (48%), Gaps = 11/165 (6%)
Frame = +1
Query: 25 DKDNKRRWSFGKQSSKTKSLPQPPPSAFNQFDSSTPLERNKHAIAVAAATAA------VA 78
+ ++KR WSF K+S++ + L N + P NK + V
Sbjct: 142 ESNDKRGWSFRKKSARHRVLS-------NTVIAEAPSSANKETSECSTFNFQPLPEPNVV 300
Query: 79 EAALAAAHAAAEVVRLTS--SGVAGSSNKTRGQLRL---PEETAAVKIQSAFRGYLARRA 133
E + + + E +L+S S +N + +L P E+ + IQ+A RG LA+R
Sbjct: 301 EK-IYTTNCSDEKPQLSSFESSQVEETNVIETEEKLDVNPPESDVIIIQAAIRGLLAQRE 477
Query: 134 LRALKALVKLQALVRGHIVRKKTADMLRRMQTLVRLQTKARASRA 178
L LK +VKLQA VRGH+VR+ LR +Q ++++Q RA RA
Sbjct: 478 LLQLKKVVKLQAAVRGHLVRRHAVGTLRXIQAIIKMQILVRARRA 612
>TC234741 weakly similar to UP|Q93ZH7 (Q93ZH7) AT5g03040/F15A17_70, partial
(10%)
Length = 423
Score = 63.9 bits (154), Expect = 1e-10
Identities = 41/111 (36%), Positives = 66/111 (58%)
Frame = +2
Query: 45 PQPPPSAFNQFDSSTPLERNKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSSGVAGSSN 104
P+ P ++ +++ E+++ A ++ ATA A AA+AA EV LT N
Sbjct: 134 PRKDPKPISEAENN---EQSRQAFSLVLATAVAAGAAVAA-----EVACLT--------N 265
Query: 105 KTRGQLRLPEETAAVKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKK 155
R + +E AA+KIQ+A+RGYLARR+LR L+ L +L+ LV+G V+++
Sbjct: 266 TPRSNGKANQEMAAIKIQTAYRGYLARRSLRGLRGLSRLKTLVQGQSVQRQ 418
>TC221116 similar to UP|Q9LK76 (Q9LK76) Arabidopsis thaliana genomic DNA,
chromosome 3, P1 clone: MDC8, partial (17%)
Length = 704
Score = 63.5 bits (153), Expect = 2e-10
Identities = 46/133 (34%), Positives = 71/133 (52%), Gaps = 8/133 (6%)
Frame = +1
Query: 268 KSDKILEVDTWKPQLNKNENNVNSMSN--ESPSKHSTKAQNQS-----LSVKFHKAKEEV 320
+S KI+E+DT+K + +++ ++MS E S H+ + +SV + ++
Sbjct: 13 ESPKIVEIDTYKTR-SRSRRFTSTMSECGEDMSCHAISSPLPCPVPGRISVPDCRHIQDF 189
Query: 321 AASRTADNSP-QTFSASSRNGSGVRRNTPFTPTRSECSWSFLGGYSGYPNYMANTESSRA 379
D T ++ R + VR N P TP +S C +F YS +PNYMANT+S A
Sbjct: 190 DWYYNVDECRFSTAHSTPRFTNYVRPNAPATPAKSVCGDTFFRPYSNFPNYMANTQSFNA 369
Query: 380 KVRSQSAPRQRHE 392
K+RS SAP+QR E
Sbjct: 370 KLRSHSAPKQRPE 408
>BF425129
Length = 415
Score = 62.4 bits (150), Expect = 4e-10
Identities = 37/79 (46%), Positives = 48/79 (59%), Gaps = 1/79 (1%)
Frame = +2
Query: 18 DGSGKKSDKDNKRRWSFGKQSSKT-KSLPQPPPSAFNQFDSSTPLERNKHAIAVAAATAA 76
D + K+ +R+W FG+ SK S+ P PS S E++KHA+ VA A+AA
Sbjct: 95 DTHSTQDKKEKRRKWIFGRLKSKRIPSIKAPLPSK-ETILSEAEEEQSKHALTVAIASAA 271
Query: 77 VAEAALAAAHAAAEVVRLT 95
AEAA+ AHAAAEVVRLT
Sbjct: 272 AAEAAVTVAHAAAEVVRLT 328
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.310 0.123 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,038,205
Number of Sequences: 63676
Number of extensions: 262757
Number of successful extensions: 1974
Number of sequences better than 10.0: 127
Number of HSP's better than 10.0 without gapping: 1806
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1910
length of query: 441
length of database: 12,639,632
effective HSP length: 100
effective length of query: 341
effective length of database: 6,272,032
effective search space: 2138762912
effective search space used: 2138762912
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC149494.9


