

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149494.2 - phase: 0
(268 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
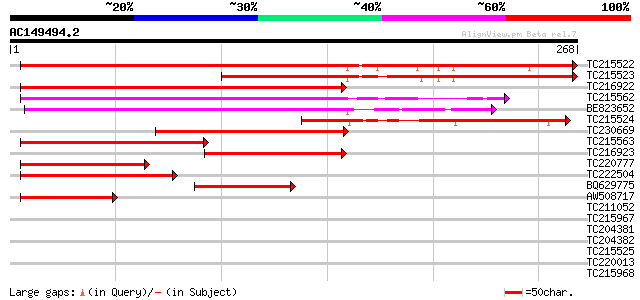
Score E
Sequences producing significant alignments: (bits) Value
TC215522 343 4e-95
TC215523 198 2e-51
TC216922 weakly similar to GB|AAL16179.1|16226483|AF428411 AT5g0... 154 4e-38
TC215562 weakly similar to UP|Q8RX71 (Q8RX71) At3g51780/ORF3, pa... 130 5e-31
BE823652 weakly similar to GP|19699283|gb| At3g51780/ORF3 {Arabi... 125 2e-29
TC215524 107 6e-24
TC230669 weakly similar to UP|Q8RX71 (Q8RX71) At3g51780/ORF3, pa... 92 2e-19
TC215563 weakly similar to UP|Q8RX71 (Q8RX71) At3g51780/ORF3, pa... 68 4e-12
TC216923 65 2e-11
TC220777 similar to GB|AAS88766.1|46359805|BT012376 At5g14360 {A... 57 7e-09
TC222504 similar to UP|Q6K5J0 (Q6K5J0) Ubiquitin-like protein, p... 57 9e-09
BQ629775 similar to GP|20160649|dbj P0446G04.18 {Oryza sativa (j... 49 3e-06
AW508717 48 5e-06
TC211052 37 0.009
TC215967 weakly similar to GB|AAP13408.1|30023750|BT006300 At5g2... 34 0.079
TC204381 similar to UP|CRTC_RICCO (P93508) Calreticulin precurso... 33 0.13
TC204382 weakly similar to UP|CRTC_RICCO (P93508) Calreticulin p... 33 0.13
TC215525 32 0.39
TC220013 similar to UP|O81998 (O81998) Transcription factor VSF-... 31 0.51
TC215968 similar to UP|Q9LLM3 (Q9LLM3) MTD1, partial (19%) 31 0.67
>TC215522
Length = 1778
Score = 343 bits (881), Expect = 4e-95
Identities = 193/278 (69%), Positives = 221/278 (79%), Gaps = 15/278 (5%)
Frame = +3
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
GELKKML+GPTGLHH+DQK+ YK+KERDSKAFLD+VGVKDKSK+V++EDPI+QEKR LE
Sbjct: 696 GELKKMLSGPTGLHHEDQKLLYKDKERDSKAFLDMVGVKDKSKIVLVEDPISQEKRLLER 875
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
RKN KME+AAKSISEISLEVDRLAG+VSA E+IISKGGKVVETDVL+LIE LMNQLLKLD
Sbjct: 876 RKNAKMEKAAKSISEISLEVDRLAGRVSAFESIISKGGKVVETDVLNLIELLMNQLLKLD 1055
Query: 126 GIVADGDVKLQRKMQVKRVQKYVETLDMLKIKN----SNGGHVPKKKPQQK----VKLPP 177
GI+ADGDVKLQRKMQV+RVQKYVETLD+LK+KN SNG P KP QK + P
Sbjct: 1056GIMADGDVKLQRKMQVRRVQKYVETLDVLKVKNSMPSSNGDQAP-VKPHQKHSNGQRKGP 1232
Query: 178 IDEQLEGMSIGNHK--LQPSLEQQSQ--RNSNGNS-QVFQQLQHKPSTNSTSEVVVTTKW 232
I EQ + S G H+ L P EQQ + RNS+ NS ++Q+ Q +PS NSTS VVVTT W
Sbjct: 1233IQEQQQKHSNGRHRLALAPIQEQQQEQPRNSSENSLALYQEQQEQPSRNSTSGVVVTTNW 1412
Query: 233 ETFDSLPPLIPV--TSASSSSSSTNNSVHPKFKWEHFE 268
E FDS+PPLIPV TS SS TNNS PKF WE F+
Sbjct: 1413ELFDSVPPLIPVQATSPPPPSSVTNNSGPPKFNWEFFD 1526
>TC215523
Length = 790
Score = 198 bits (503), Expect = 2e-51
Identities = 114/177 (64%), Positives = 133/177 (74%), Gaps = 9/177 (5%)
Frame = +2
Query: 101 KGGKVVETDVLSLIEKLMNQLLKLDGIVADGDVKLQRKMQVKRVQKYVETLDMLKIKN-- 158
KGG+VVETD+L+LIE LMNQLLKLDGI+ADGDVKLQRKMQVKRVQKYVETLD+LK+KN
Sbjct: 2 KGGRVVETDLLNLIELLMNQLLKLDGIMADGDVKLQRKMQVKRVQKYVETLDVLKVKNSM 181
Query: 159 --SNGGHVPKKKPQQKVKLPPIDEQLEGMSIGNHKLQ--PSLEQQSQ--RNSNGNS-QVF 211
SNG H P +PQQK + Q + S G+H+L P EQQ + RNSN NS +++
Sbjct: 182 PSSNGDHAP-VQPQQKHS----NGQQQKYSNGHHRLALVPIQEQQQEQPRNSNENSLELY 346
Query: 212 QQLQHKPSTNSTSEVVVTTKWETFDSLPPLIPVTSASSSSSSTNNSVHPKFKWEHFE 268
Q+ QH+PS NSTS VVVTT WE FDS+PPLIPV S S SS TNNS PKF WE F+
Sbjct: 347 QEQQHQPSRNSTSGVVVTTNWELFDSVPPLIPVQSTSPPSSVTNNSGPPKFNWEFFD 517
>TC216922 weakly similar to GB|AAL16179.1|16226483|AF428411
AT5g07220/T28J14_160 {Arabidopsis thaliana;} , partial
(15%)
Length = 1055
Score = 154 bits (389), Expect = 4e-38
Identities = 73/154 (47%), Positives = 115/154 (74%)
Frame = +1
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
GE+KK+L+G TGL +Q++ Y+ KER++ +LD+ GVKD+SK+V+++DP + E+R+++M
Sbjct: 196 GEVKKVLSGETGLEVDEQRLLYRGKERENGEYLDVCGVKDRSKVVLIQDPSSIERRFIQM 375
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
R N K++ A ++I+ +++++D+LA QVSA+E IS G KV E + +LIE LM Q +KL+
Sbjct: 376 RINAKIQTAHRAINNVAVQLDQLADQVSAIEKSISNGVKVPEVQITTLIEMLMRQAIKLE 555
Query: 126 GIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNS 159
I A+GD Q+ +Q KRVQK VE LD LK+ N+
Sbjct: 556 SISAEGDASAQKNLQGKRVQKCVEKLDQLKVSNA 657
>TC215562 weakly similar to UP|Q8RX71 (Q8RX71) At3g51780/ORF3, partial (64%)
Length = 1128
Score = 130 bits (328), Expect = 5e-31
Identities = 84/231 (36%), Positives = 124/231 (53%)
Frame = +2
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
G++KK+L TGL +Q++F++ E+ L GVKDKSKL ++E ++E++ E
Sbjct: 260 GDVKKLLVNKTGLEPAEQRLFFRGIEKGDNQRLQAEGVKDKSKLFLLEGIGSKERKLEET 439
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLD 125
RK +M +A ++I+ + EVD+L+ +V+++E I+ G K E + L L E LM+QLLKL+
Sbjct: 440 RKENEMSKAFEAIASVRAEVDKLSNRVTSIEVAINGGNKASEKEFLVLTELLMSQLLKLN 619
Query: 126 GIVADGDVKLQRKMQVKRVQKYVETLDMLKIKNSNGGHVPKKKPQQKVKLPPIDEQLEGM 185
GI A+G+ KLQRK +V RVQ V+ LD LK +N+N P VK + Q E
Sbjct: 620 GIEAEGEAKLQRKAEVNRVQNLVDKLDSLKARNAN----PFSNSSNAVK---VTTQWETF 778
Query: 186 SIGNHKLQPSLEQQSQRNSNGNSQVFQQLQHKPSTNSTSEVVVTTKWETFD 236
G L PS N +S VT +WE FD
Sbjct: 779 DSGMESLD-----------------------APSDNPSS-TKVTQEWERFD 859
>BE823652 weakly similar to GP|19699283|gb| At3g51780/ORF3 {Arabidopsis
thaliana}, partial (31%)
Length = 742
Score = 125 bits (315), Expect = 2e-29
Identities = 81/227 (35%), Positives = 127/227 (55%), Gaps = 4/227 (1%)
Frame = -3
Query: 8 LKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEMRK 67
LK T +Q+ ++ KE++ + L +VGVKD K++++EDP ++E++ EM K
Sbjct: 737 LKXXXTSEXXXXXXEQRXLFRGKEKEDEECLHMVGVKDMXKVILLEDPASKERKLEEMHK 558
Query: 68 NIKMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLDGI 127
+ + +A ++IS++ EVD+L +V ALET + G KV + + L E LM QLLKLD I
Sbjct: 557 SEDISKACEAISKVRTEVDKLYQKVVALETTVCGGAKVEDKEFAILTELLMVQLLKLDSI 378
Query: 128 VADGDVKLQRKMQVKRVQKYVETLDMLKIKN----SNGGHVPKKKPQQKVKLPPIDEQLE 183
A+G+ K QR+++V+RVQ YV+T+D LK +N SN G+ +P I E LE
Sbjct: 377 AAEGEAKGQRRVEVRRVQSYVDTIDNLKARNFAAFSNAGN---------NAVPVIWEALE 225
Query: 184 GMSIGNHKLQPSLEQQSQRNSNGNSQVFQQLQHKPSTNSTSEVVVTT 230
+G+ K S + N + F+ +Q PST E + T
Sbjct: 224 S-GVGSLKSPTSFPTSTVITQNW--EHFE*IQVNPSTLFIEEASMMT 93
>TC215524
Length = 716
Score = 107 bits (267), Expect = 6e-24
Identities = 69/138 (50%), Positives = 86/138 (62%), Gaps = 11/138 (7%)
Frame = +1
Query: 139 MQVKRVQKYVETLDMLKIKNS----NGGHVPKKKPQQKVKLPPIDEQLEGMSIGNHKLQP 194
++VKRVQKYVETLD+LK+KNS NGGH P + PQQK + Q +G P
Sbjct: 82 LKVKRVQKYVETLDVLKVKNSMPSSNGGHAPVQ-PQQKHS----NGQRQG---------P 219
Query: 195 SLEQQSQRNSNGNSQ-----VFQQLQHKPSTNSTSEVVVTTKWETFDSLPPLIPVTSASS 249
E++ Q++SNG+ + + +Q Q +PS NSTS VVVTT WE FDS+PPLIPV S S
Sbjct: 220 VQEKKQQKHSNGHHRLALAPIQEQEQQQPSRNSTSGVVVTTNWELFDSVPPLIPVPSTSP 399
Query: 250 SSSS--TNNSVHPKFKWE 265
S TNNS KF WE
Sbjct: 400 SPHPPVTNNSGPHKFNWE 453
>TC230669 weakly similar to UP|Q8RX71 (Q8RX71) At3g51780/ORF3, partial (49%)
Length = 674
Score = 92.4 bits (228), Expect = 2e-19
Identities = 47/91 (51%), Positives = 66/91 (71%)
Frame = +1
Query: 70 KMERAAKSISEISLEVDRLAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLDGIVA 129
+M A+++++E+ EVD+LA +VS LE + G +V + + L E LM QLLKLD I A
Sbjct: 4 EMLEASEAVAEVRAEVDKLAERVSVLEVAVDGGTRVSDKEFLMSTELLMRQLLKLDSIEA 183
Query: 130 DGDVKLQRKMQVKRVQKYVETLDMLKIKNSN 160
+G+VKLQRK +V+RVQ +V+TLD LK KNSN
Sbjct: 184 EGEVKLQRKAEVRRVQNFVDTLDSLKAKNSN 276
Score = 34.3 bits (77), Expect = 0.060
Identities = 34/116 (29%), Positives = 48/116 (41%), Gaps = 9/116 (7%)
Frame = +1
Query: 144 VQKYVETLDMLKIKNSNGGHVPKKK--------PQQKVKLPPIDEQLEGMSIGNHKLQPS 195
V K E + +L++ G V K+ +Q +KL I+ + G KLQ
Sbjct: 49 VDKLAERVSVLEVAVDGGTRVSDKEFLMSTELLMRQLLKLDSIEAE------GEVKLQRK 210
Query: 196 LEQQSQRNSNGNSQVFQQLQHKPSTNSTSEVVVTTKWETFDS-LPPLIPVTSASSS 250
E + +N + P T V V T+WETFDS + L TS SSS
Sbjct: 211 AEVRRVQNFVDTLDSLKAKNSNPFTPIGKAVSVATQWETFDSGMGSLNAPTSMSSS 378
>TC215563 weakly similar to UP|Q8RX71 (Q8RX71) At3g51780/ORF3, partial (18%)
Length = 495
Score = 68.2 bits (165), Expect = 4e-12
Identities = 32/89 (35%), Positives = 59/89 (65%)
Frame = +2
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
G++KK+L TGL +Q++F++ E+ L + GVKDKSKL+++E ++E++ E
Sbjct: 227 GDVKKLLVNKTGLEPVEQRLFFRGIEKGDNLHLHLEGVKDKSKLLLLEGTASKERKLEET 406
Query: 66 RKNIKMERAAKSISEISLEVDRLAGQVSA 94
RK M +A ++I+ + EVD+L+ +V++
Sbjct: 407 RKQNVMSKAFEAIAGVRAEVDKLSXRVTS 493
>TC216923
Length = 629
Score = 65.5 bits (158), Expect = 2e-11
Identities = 34/67 (50%), Positives = 44/67 (64%)
Frame = +2
Query: 93 SALETIISKGGKVVETDVLSLIEKLMNQLLKLDGIVADGDVKLQRKMQVKRVQKYVETLD 152
SA+E IS G KV E + +LIE LM Q +KL+ I A+G Q+ +Q KRVQK VE LD
Sbjct: 2 SAIEKSISNGVKVPEVQITTLIEMLMRQAIKLESISAEGGASAQKNLQGKRVQKCVEKLD 181
Query: 153 MLKIKNS 159
LK+ N+
Sbjct: 182 QLKVSNA 202
>TC220777 similar to GB|AAS88766.1|46359805|BT012376 At5g14360 {Arabidopsis
thaliana;} , partial (64%)
Length = 735
Score = 57.4 bits (137), Expect = 7e-09
Identities = 25/61 (40%), Positives = 42/61 (67%)
Frame = +1
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
GELK +L+ T ++Q++ +K KER+ +L +VGV+DK K+++ EDP +EK+ L +
Sbjct: 307 GELKMILSLATSFEPREQRLLFKGKEREDDEYLHMVGVRDKDKVLLFEDPAIKEKKLLGL 486
Query: 66 R 66
R
Sbjct: 487 R 489
>TC222504 similar to UP|Q6K5J0 (Q6K5J0) Ubiquitin-like protein, partial (60%)
Length = 774
Score = 57.0 bits (136), Expect = 9e-09
Identities = 26/74 (35%), Positives = 47/74 (63%)
Frame = +2
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEM 65
GELK +L+ T L ++Q++ ++ KE++ FL +VGV+DK K++++EDP +E + L M
Sbjct: 317 GELKMILSLVTSLEPREQRLLFRGKEKEDNEFLHMVGVRDKDKVLLLEDPAIKEMKLLGM 496
Query: 66 RKNIKMERAAKSIS 79
+ + +IS
Sbjct: 497 ARGQPINNTCCTIS 538
>BQ629775 similar to GP|20160649|dbj P0446G04.18 {Oryza sativa (japonica
cultivar-group)}, partial (9%)
Length = 435
Score = 48.5 bits (114), Expect = 3e-06
Identities = 25/48 (52%), Positives = 33/48 (68%)
Frame = +2
Query: 88 LAGQVSALETIISKGGKVVETDVLSLIEKLMNQLLKLDGIVADGDVKL 135
L G V+++E I+ G K E + L L E LM+QLLKLDGI A+G+ KL
Sbjct: 290 LRGSVTSIEVSINGGNKASEKEFLVLTELLMSQLLKLDGIEAEGEAKL 433
>AW508717
Length = 288
Score = 47.8 bits (112), Expect = 5e-06
Identities = 21/46 (45%), Positives = 32/46 (68%)
Frame = +1
Query: 6 GELKKMLTGPTGLHHQDQKIFYKNKERDSKAFLDIVGVKDKSKLVV 51
G LK +LT TGL ++Q++ ++ KERD L +VGVKD SK+++
Sbjct: 151 GHLKMVLTSETGLEPKEQRLLFRGKERDDDECLHMVGVKDMSKVIL 288
>TC211052
Length = 635
Score = 37.0 bits (84), Expect = 0.009
Identities = 19/51 (37%), Positives = 29/51 (56%)
Frame = +2
Query: 218 PSTNSTSEVVVTTKWETFDSLPPLIPVTSASSSSSSTNNSVHPKFKWEHFE 268
P +NS+++V VTT+WETFDS +S + S ++S WE F+
Sbjct: 287 PFSNSSNDVKVTTQWETFDS------GMESSDAPSDNSSSTKVTQDWEQFD 421
>TC215967 weakly similar to GB|AAP13408.1|30023750|BT006300 At5g21940
{Arabidopsis thaliana;} , partial (26%)
Length = 1034
Score = 33.9 bits (76), Expect = 0.079
Identities = 21/79 (26%), Positives = 35/79 (43%), Gaps = 1/79 (1%)
Frame = +2
Query: 26 FYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEMR-KNIKMERAAKSISEISLE 84
FY K + + D ++V EDP A++++ L R +I+ R+ +I IS
Sbjct: 344 FYSGKSKSFTSLADAAAASSMEEIVKPEDPYAKKRKNLIARNSSIERSRSCANIGGISKR 523
Query: 85 VDRLAGQVSALETIISKGG 103
+ G S L S+ G
Sbjct: 524 PSNIGGGASCLTLNCSEEG 580
>TC204381 similar to UP|CRTC_RICCO (P93508) Calreticulin precursor, partial
(81%)
Length = 1408
Score = 33.1 bits (74), Expect = 0.13
Identities = 19/49 (38%), Positives = 27/49 (54%)
Frame = -3
Query: 214 LQHKPSTNSTSEVVVTTKWETFDSLPPLIPVTSASSSSSSTNNSVHPKF 262
LQ S++ T +V++ FDS +P +SASSSSSS + S F
Sbjct: 1055 LQTS*SSSCTPSSLVSSPASVFDSASESLPASSASSSSSSASESTGSSF 909
>TC204382 weakly similar to UP|CRTC_RICCO (P93508) Calreticulin precursor,
partial (12%)
Length = 449
Score = 33.1 bits (74), Expect = 0.13
Identities = 19/49 (38%), Positives = 27/49 (54%)
Frame = -1
Query: 214 LQHKPSTNSTSEVVVTTKWETFDSLPPLIPVTSASSSSSSTNNSVHPKF 262
LQ S++ T +V++ FDS +P +SASSSSSS + S F
Sbjct: 197 LQTS*SSSCTPSSLVSSPASVFDSASESLPASSASSSSSSASESTGSSF 51
>TC215525
Length = 395
Score = 31.6 bits (70), Expect = 0.39
Identities = 14/26 (53%), Positives = 15/26 (56%)
Frame = +1
Query: 242 IPVTSASSSSSSTNNSVHPKFKWEHF 267
+P TS S S TN S PKF WE F
Sbjct: 1 VPSTSPSPHPSETNKSGPPKFNWEFF 78
>TC220013 similar to UP|O81998 (O81998) Transcription factor VSF-1, partial
(29%)
Length = 1572
Score = 31.2 bits (69), Expect = 0.51
Identities = 36/116 (31%), Positives = 56/116 (48%), Gaps = 5/116 (4%)
Frame = +3
Query: 148 VETLDMLKIKNSNGGHVP-KKKPQQKVKLPPIDEQLEGMSIGNHKLQPSLEQQSQRNSNG 206
VE+++ + +NS+ VP + P QK +PP S N + PS S+ +
Sbjct: 24 VESMEGVDQRNSSAFGVPVPRPPNQKHGIPP--------SHPNINISPSATPCSRYS--- 170
Query: 207 NSQVFQQLQHKPSTNSTSEVVVTTKWETFDSLPPLIP----VTSASSSSSSTNNSV 258
Q+L+ S++S S + + + DSLPPL P VTS S S+T+ SV
Sbjct: 171 -----QRLRSPGSSHSRS--LSQSPIFSLDSLPPLSPSPSAVTSMFDSISTTDMSV 317
>TC215968 similar to UP|Q9LLM3 (Q9LLM3) MTD1, partial (19%)
Length = 942
Score = 30.8 bits (68), Expect = 0.67
Identities = 22/78 (28%), Positives = 37/78 (47%)
Frame = +1
Query: 26 FYKNKERDSKAFLDIVGVKDKSKLVVMEDPIAQEKRYLEMRKNIKMERAAKSISEISLEV 85
FY K R + D ++V EDP A++++ L + +N +ER ++S + I
Sbjct: 340 FYSGKSRSFTSLADAAAASSMEEIVKPEDPYAKKRKNL-IARNSSIER-SRSCANI---- 501
Query: 86 DRLAGQVSALETIISKGG 103
G +S T I +GG
Sbjct: 502 ----GGISKRPTNIGRGG 543
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.311 0.129 0.350
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,624,895
Number of Sequences: 63676
Number of extensions: 117124
Number of successful extensions: 1058
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 997
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1039
length of query: 268
length of database: 12,639,632
effective HSP length: 96
effective length of query: 172
effective length of database: 6,526,736
effective search space: 1122598592
effective search space used: 1122598592
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC149494.2


