

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149490.9 - phase: 0
(1049 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
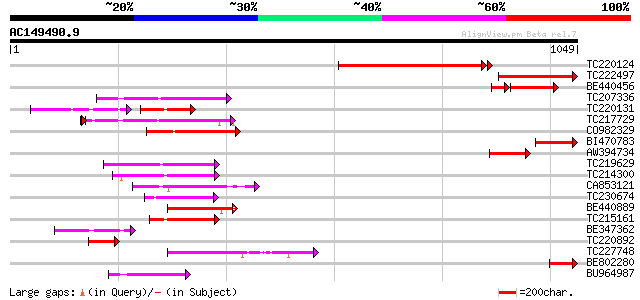
Score E
Sequences producing significant alignments: (bits) Value
TC220124 weakly similar to UP|Q9LZU5 (Q9LZU5) Kinesin-related pr... 482 e-135
TC222497 similar to UP|Q94G20 (Q94G20) KRP120-2, partial (13%) 256 5e-68
BE440456 similar to GP|15186760|gb KRP120-2 {Daucus carota}, par... 167 2e-51
TC207336 similar to UP|O24147 (O24147) Kinesin-like protein, par... 160 3e-39
TC220131 similar to UP|ATK2_ARATH (P46864) Kinesin 2 (Kinesin-li... 104 3e-38
TC217729 similar to UP|Q9CAC9 (Q9CAC9) Kinesin-like protein; 736... 149 2e-36
CO982329 134 2e-31
BI470783 similar to GP|15186760|gb KRP120-2 {Daucus carota}, par... 131 1e-30
AW394734 similar to GP|15186760|gb KRP120-2 {Daucus carota}, par... 130 3e-30
TC219629 similar to UP|ATK1_ARATH (Q07970) Kinesin 1 (Kinesin-li... 125 7e-29
TC214300 similar to UP|ATK1_ARATH (Q07970) Kinesin 1 (Kinesin-li... 121 2e-27
CA853121 114 2e-25
TC230674 homologue to UP|Q9M0X6 (Q9M0X6) Kinesin-like protein, p... 101 2e-21
BE440889 100 3e-21
TC215161 similar to UP|Q6NQ77 (Q6NQ77) At4g05190, partial (19%) 98 2e-20
BE347362 96 1e-19
TC220892 homologue to UP|K125_TOBAC (O23826) 125 kDa kinesin-rel... 91 3e-18
TC227748 similar to UP|Q940Y8 (Q940Y8) AT3g16060/MSL1_10, partia... 89 8e-18
BE802280 similar to GP|15186760|gb KRP120-2 {Daucus carota}, par... 86 7e-17
BU964987 similar to GP|15208447|gb kinesin heavy chain {Zea mays... 82 9e-16
>TC220124 weakly similar to UP|Q9LZU5 (Q9LZU5) Kinesin-related protein-like,
partial (26%)
Length = 864
Score = 482 bits (1240), Expect(2) = e-135
Identities = 246/275 (89%), Positives = 265/275 (95%)
Frame = +3
Query: 608 SFVSTKSEATEDLRVRVVELKNMYGSGIKALDNLAEELKSNNQLTYEDLKSEVAKHSSAL 667
SFVSTK+EATEDLR RV +LKNMYGSGIKALD+LAEELK NNQLTY+DLKSEVAKHSSAL
Sbjct: 3 SFVSTKAEATEDLRQRVGKLKNMYGSGIKALDDLAEELKVNNQLTYDDLKSEVAKHSSAL 182
Query: 668 EDLFKGIALEADSLLNDLQNSLHKQEANVTAFAHQQREAHSRAVETTRSVSKITMKFFET 727
EDLFKGIALEADSLLNDLQ+SLHKQEAN+TA+AHQQRE+H+RAVETTR+VSKIT+ FFET
Sbjct: 183 EDLFKGIALEADSLLNDLQSSLHKQEANLTAYAHQQRESHARAVETTRAVSKITVNFFET 362
Query: 728 IDRHASSLTQIVEETQFVNDQKLCELEKKFEECTAYEEKQLLEKVAEMLASSNARKKKLV 787
IDRHASSLT+IVEE Q VNDQKLCELEKKFEECTAYEEKQLLEKVAEMLASSNARKK+LV
Sbjct: 363 IDRHASSLTEIVEEAQLVNDQKLCELEKKFEECTAYEEKQLLEKVAEMLASSNARKKQLV 542
Query: 788 QMAVNDLRESANCRTSKLQREALTMQDSTSFVKAEWMVHMEKTESNYHEDTSSVESGKKD 847
QMAVNDLRESANCRTSKL++EALTMQDSTS VKAEW VHMEKTESNYHEDTS+VESGK+D
Sbjct: 543 QMAVNDLRESANCRTSKLRQEALTMQDSTSSVKAEWRVHMEKTESNYHEDTSAVESGKRD 722
Query: 848 LAEVLQICLNKAEVGSQQWRNAQDSLLSLEKRNAG 882
L EVLQICLNKA+VGSQQWR AQ+SLLSLEKRNAG
Sbjct: 723 LVEVLQICLNKAKVGSQQWRKAQESLLSLEKRNAG 827
Score = 21.6 bits (44), Expect(2) = e-135
Identities = 9/10 (90%), Positives = 9/10 (90%)
Frame = +2
Query: 884 VDTIVRGGME 893
VDTIV GGME
Sbjct: 833 VDTIVTGGME 862
>TC222497 similar to UP|Q94G20 (Q94G20) KRP120-2, partial (13%)
Length = 706
Score = 256 bits (653), Expect = 5e-68
Identities = 124/146 (84%), Positives = 133/146 (90%)
Frame = +1
Query: 904 SSVSTTLEDAGIANTDINSSIDYSLQLDHEACGNLNSMITPCCGDLTELKGGHYNRIVEI 963
S+VS TLEDAGIA DINSSID+SLQLDHEACGNLNSMI PCCGDL ELKGGH++ IVEI
Sbjct: 1 SAVSATLEDAGIAXKDINSSIDHSLQLDHEACGNLNSMIIPCCGDLRELKGGHFHSIVEI 180
Query: 964 TENAGKCLLNEYMVDEPSCSTPTRRLFNLPSVSSIEELRTPSFEELLKAFWDAKSQKLAN 1023
TEN+GKCLLNEYMVDEPSCSTP +RLFNLP VSSIEELRTPSFEELLK+FWDA+S K AN
Sbjct: 181 TENSGKCLLNEYMVDEPSCSTPRKRLFNLPCVSSIEELRTPSFEELLKSFWDARSPKQAN 360
Query: 1024 GDVKHIGSYEETQSVRDSRVPLTTIN 1049
GDVKHIG+YE QSVRDSRVPLT IN
Sbjct: 361 GDVKHIGAYEAAQSVRDSRVPLTAIN 438
>BE440456 similar to GP|15186760|gb KRP120-2 {Daucus carota}, partial (8%)
Length = 464
Score = 167 bits (424), Expect(2) = 2e-51
Identities = 78/88 (88%), Positives = 83/88 (93%)
Frame = +1
Query: 927 SLQLDHEACGNLNSMITPCCGDLTELKGGHYNRIVEITENAGKCLLNEYMVDEPSCSTPT 986
SLQLDH+ACGNLNSMI PCCGDL ELKGGHY+RIVEITENAGKCLL+EY VDEPSCSTP
Sbjct: 199 SLQLDHDACGNLNSMIIPCCGDLRELKGGHYHRIVEITENAGKCLLSEYTVDEPSCSTPR 378
Query: 987 RRLFNLPSVSSIEELRTPSFEELLKAFW 1014
+R FNLPSVSSIEELRTPSFEELLK+FW
Sbjct: 379 KRSFNLPSVSSIEELRTPSFEELLKSFW 462
Score = 54.7 bits (130), Expect(2) = 2e-51
Identities = 28/35 (80%), Positives = 30/35 (85%)
Frame = +2
Query: 891 GMEANQALRARFSSSVSTTLEDAGIANTDINSSID 925
GMEAN LR RFSS+VSTTLEDA IAN DINSSI+
Sbjct: 2 GMEANHLLRDRFSSAVSTTLEDAEIANKDINSSIE 106
>TC207336 similar to UP|O24147 (O24147) Kinesin-like protein, partial (22%)
Length = 1055
Score = 160 bits (405), Expect = 3e-39
Identities = 98/254 (38%), Positives = 144/254 (56%), Gaps = 4/254 (1%)
Frame = +3
Query: 161 GVIPRAVKQIFDILEAQSAEYS--MKVTFLELYNEEITDLLAPEETTKFVDEKSKKPIAL 218
G+ PRA+ ++F IL + +YS +K +ELY + + DLL P K+ KP+ L
Sbjct: 9 GLTPRAIAELFRILRRDNNKYSFSLKAYMVELYQDTLIDLLLP---------KNGKPLKL 161
Query: 219 --MEDGKGGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIH 276
+D G V+V + + T E+ I+++GS +R + T +N +SSRSH I SI I
Sbjct: 162 DIKKDSTGMVVVENVTVMSISTIEELNSIIQRGSERRHISGTQMNDESSRSHLILSIVI- 338
Query: 277 IKECTPEGEEMIKCGKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINALVE 336
E T + + GKL+ VDLAGSE + +SG+ + +EA INKSL LG I++L
Sbjct: 339 --ESTNLQSQSVAKGKLSFVDLAGSERVKKSGSTGSQLKEAQSINKSLSALGDVISSLSS 512
Query: 337 HSGHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPE 396
H PYR+ KLT L+ DSLGG KT + V+P+ L+ET ++L YA R ++I N P
Sbjct: 513 GGQHTPYRNHKLTMLMSDSLGGNAKTLMFVNVAPTESNLDETNNSLMYASRVRSIVNDPN 692
Query: 397 VNQKMMKSAMIKDL 410
N + A +K L
Sbjct: 693 KNVSSKEVARLKKL 734
>TC220131 similar to UP|ATK2_ARATH (P46864) Kinesin 2 (Kinesin-like protein
B), partial (37%)
Length = 1044
Score = 104 bits (259), Expect(2) = 3e-38
Identities = 67/189 (35%), Positives = 98/189 (51%), Gaps = 3/189 (1%)
Frame = +3
Query: 39 HNKYDKEKGVNVQVLVRCRPMNEDEMRLHTPVVISCNEGRREVAAVQSIANKQIDRTFVF 98
HN + KG N++V R RP+ DE + S +A F F
Sbjct: 150 HNTILELKG-NIRVFCRVRPLLADESCSTEGRIFSYPTSMETSGRAIDLAQNGQKHAFTF 326
Query: 99 DKVFGPNSQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTYTMEGGAIKKNGEFPT 158
DKVF P + Q+E++ + +S +V L+GY IFAYGQTG+GKTYTM G P
Sbjct: 327 DKVFTPEASQEEVFVE-ISQLVQSALDGYKVCIFAYGQTGSGKTYTMMG-----RPGHPE 488
Query: 159 DAGVIPRAVKQIFDILEAQSAE---YSMKVTFLELYNEEITDLLAPEETTKFVDEKSKKP 215
+ G+IPR+++QIF ++Q + Y M+V+ LE+YNE I DL++ TT+ + K
Sbjct: 489 EKGLIPRSLEQIFQTKQSQQPQGWKYEMQVSMLEIYNETIRDLIS--TTTRMENGTPGKQ 662
Query: 216 IALMEDGKG 224
+ D G
Sbjct: 663 YTIKHDTNG 689
Score = 73.9 bits (180), Expect(2) = 3e-38
Identities = 41/103 (39%), Positives = 64/103 (61%), Gaps = 1/103 (0%)
Frame = +1
Query: 242 IYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIH-IKECTPEGEEMIKCGKLNLVDLAG 300
++ +L + + R +T +N+QSSRSH +F++ I+ + E T + + G LNL+DLAG
Sbjct: 748 LHFLLNQAANSRSVGKTQMNEQSSRSHFVFTLRIYGVNESTDQQVQ----GVLNLIDLAG 915
Query: 301 SENISRSGAREGRAREAGEINKSLLTLGRTINALVEHSGHVPY 343
SE +S+SG+ R +E INKSL +L I AL + HVP+
Sbjct: 916 SERLSKSGSTGDRLKETQAINKSLSSLSDVIFALAKKEDHVPF 1044
>TC217729 similar to UP|Q9CAC9 (Q9CAC9) Kinesin-like protein; 73641-79546,
partial (26%)
Length = 1075
Score = 149 bits (375), Expect(2) = 2e-36
Identities = 103/293 (35%), Positives = 162/293 (55%), Gaps = 15/293 (5%)
Frame = +1
Query: 141 KTYTMEGGAIKKNGEFPTDAGVIPRAVKQIFDILEAQ--SAEYSMKVTFLELYNEEITDL 198
KTYTM G + +D GV RA+ +F I +++ S Y + V +E+YNE++ DL
Sbjct: 37 KTYTMSGPGLSSK----SDWGVNYRALHDLFHISQSRRSSIVYEVGVQMVEIYNEQVRDL 204
Query: 199 LAPEETTKFVDEKSKKPIALMEDGK-GGVLVRGLEEEIVCTANEIYKILEKGSAKRRTAE 257
L+ +K + + + G+ V V + ++ +++ G R T+
Sbjct: 205 LS--------SNGPQKRLGIWNTAQPNGLAVPDASMHSVNSMADVLELMNIGLMNRATSA 360
Query: 258 TLLNKQSSRSHSIFSITIHIKECTPEGEEMIKCGKLNLVDLAGSENISRSGAREGRAREA 317
T LN++SSRSHS+ S+ H++ + +++ G L+LVDLAGSE + RS A +EA
Sbjct: 361 TALNERSSRSHSVLSV--HVRGTDLKTNTLLR-GCLHLVDLAGSERVDRSEATGDMLKEA 531
Query: 318 GEINKSLLTLGRTINALVEHSGHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEE 377
INKSL LG I AL + S HVPYR+SKLT+LL+ SLGG+ KT + ++P + E
Sbjct: 532 QHINKSLSALGDVIFALSQKSSHVPYRNSKLTQLLQSSLGGQAKTLMFVQLNPDVASYSE 711
Query: 378 TLSTLDYAHR--------AKNIKNKPEVNQKMMKSAMIKDLYS----EIDRLK 418
T+STL +A R A++ K +V + M + A +KD+ + EI+RL+
Sbjct: 712 TVSTLKFAERVSGVELGAARSNKEGRDVRELMEQLASLKDVIARKDEEIERLQ 870
Score = 23.1 bits (48), Expect(2) = 2e-36
Identities = 9/11 (81%), Positives = 10/11 (90%)
Frame = +3
Query: 131 IFAYGQTGTGK 141
IFAYGQTG+ K
Sbjct: 6 IFAYGQTGSXK 38
>CO982329
Length = 852
Score = 134 bits (338), Expect = 2e-31
Identities = 83/176 (47%), Positives = 108/176 (61%), Gaps = 2/176 (1%)
Frame = -3
Query: 253 RRTAETLLNKQSSRSHSIFSITIHIKECTPEGE-EMIKCGKLNLVDLAGSENISRSGARE 311
R T N SSRSH+IFS+TI C E E I +LNL+DLAGSE+ SR+
Sbjct: 832 RHVGSTNFNLLSSRSHTIFSLTIESSPCGENNEGEAITLSQLNLIDLAGSES-SRAETTG 656
Query: 312 GRAREAGEINKSLLTLGRTINALVE-HSGHVPYRDSKLTRLLRDSLGGKTKTCIIATVSP 370
R RE INKSLLTLG I+ L E + H+PYRDSKLTRLL+ SL G + +I TV+P
Sbjct: 655 MRRREGSYINKSLLTLGTVISKLTEGKASHIPYRDSKLTRLLQSSLSGHGRISLICTVTP 476
Query: 371 SIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMKSAMIKDLYSEIDRLKQEVYAARE 426
S EET +TL +AHRAK+I+ + N + + ++IK EI LK+E+ R+
Sbjct: 475 SSSNAEETHNTLKFAHRAKHIEIQAAQNTIIDEKSLIKKYQHEIQFLKEELEQMRQ 308
>BI470783 similar to GP|15186760|gb KRP120-2 {Daucus carota}, partial (5%)
Length = 430
Score = 131 bits (330), Expect = 1e-30
Identities = 64/77 (83%), Positives = 68/77 (88%)
Frame = +1
Query: 973 NEYMVDEPSCSTPTRRLFNLPSVSSIEELRTPSFEELLKAFWDAKSQKLANGDVKHIGSY 1032
NEYMVDEPSCSTP +RLFNL SVSSIEELRTPSFEELLK+FWDA+S K ANGDVKHIG+Y
Sbjct: 1 NEYMVDEPSCSTPRKRLFNLSSVSSIEELRTPSFEELLKSFWDARSPKQANGDVKHIGAY 180
Query: 1033 EETQSVRDSRVPLTTIN 1049
E VRDSRVPLT IN
Sbjct: 181 EAALFVRDSRVPLTAIN 231
>AW394734 similar to GP|15186760|gb KRP120-2 {Daucus carota}, partial (7%)
Length = 229
Score = 130 bits (327), Expect = 3e-30
Identities = 63/75 (84%), Positives = 68/75 (90%)
Frame = +3
Query: 889 RGGMEANQALRARFSSSVSTTLEDAGIANTDINSSIDYSLQLDHEACGNLNSMITPCCGD 948
RGGMEAN LR RFSS+VSTTLEDA IAN DINSSI++SLQLDH+ACGNLNSMI PCCGD
Sbjct: 3 RGGMEANHLLRDRFSSAVSTTLEDAEIANKDINSSIEHSLQLDHDACGNLNSMIIPCCGD 182
Query: 949 LTELKGGHYNRIVEI 963
L ELKGGHY+RIVEI
Sbjct: 183 LRELKGGHYHRIVEI 227
>TC219629 similar to UP|ATK1_ARATH (Q07970) Kinesin 1 (Kinesin-like protein
A), partial (26%)
Length = 923
Score = 125 bits (315), Expect = 7e-29
Identities = 80/214 (37%), Positives = 115/214 (53%)
Frame = +1
Query: 174 LEAQSAEYSMKVTFLELYNEEITDLLAPEETTKFVDEKSKKPIALMEDGKGGVLVRGLEE 233
L+ Q +Y+M V+ E+YNE I DLL+ ++ +++ L
Sbjct: 31 LKDQGWKYTMHVSIYEIYNETIRDLLSSNRSSGNDHTRTENSAPTPSKQHTIKHESDLAT 210
Query: 234 EIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPEGEEMIKCGKL 293
VC+A EI +L++ + R T +N+QSSRSH +F + I + E E G L
Sbjct: 211 LEVCSAEEISSLLQQAAQSRSVGRTQMNEQSSRSHFVFKLRISGRN---EKTEKQVQGVL 381
Query: 294 NLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINALVEHSGHVPYRDSKLTRLLR 353
NL+DLAGSE +SRSGA R +E INKSL +L I AL + HVP+R+SKLT L+
Sbjct: 382 NLIDLAGSERLSRSGATGDRLKETQAINKSLSSLSDVIFALAKKEEHVPFRNSKLTHFLQ 561
Query: 354 DSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHR 387
LGG +KT + +SP E+L +L +A R
Sbjct: 562 PYLGGDSKTLMFVNISPDQSSAGESLCSLRFAAR 663
>TC214300 similar to UP|ATK1_ARATH (Q07970) Kinesin 1 (Kinesin-like protein
A), partial (27%)
Length = 903
Score = 121 bits (303), Expect = 2e-27
Identities = 76/205 (37%), Positives = 114/205 (55%), Gaps = 7/205 (3%)
Frame = +2
Query: 190 LYNEEITDLLAPEET------TKFVDEKSKKPIALMEDGKGGVLVRGLEEEIVCTANEIY 243
+YNE I DLLA ++ T+ + K + D G V L V + E+
Sbjct: 2 IYNETIRDLLATNKSSADGTPTRVENGTPGKQYMIKHDANGNTHVSDLTVVDVQSVKEVA 181
Query: 244 KILEKGSAKRRTAETLLNKQSSRSHSIFSITIH-IKECTPEGEEMIKCGKLNLVDLAGSE 302
+L + ++ R +T +N+QSSRSH +F++ I+ + E T + + G LNL+DLAGSE
Sbjct: 182 FLLNQAASSRSVGKTQMNEQSSRSHFVFTLRIYGVNESTDQQVQ----GILNLIDLAGSE 349
Query: 303 NISRSGAREGRAREAGEINKSLLTLGRTINALVEHSGHVPYRDSKLTRLLRDSLGGKTKT 362
+SRSG+ R +E INKSL +L I AL + H+P+R+SKLT LL+ LGG +KT
Sbjct: 350 RLSRSGSTGDRLKETQAINKSLSSLSDVIFALAKKEDHIPFRNSKLTYLLQPCLGGDSKT 529
Query: 363 CIIATVSPSIHCLEETLSTLDYAHR 387
+ +SP E+L +L +A R
Sbjct: 530 LMFVNISPDQASSGESLCSLRFASR 604
>CA853121
Length = 677
Score = 114 bits (285), Expect = 2e-25
Identities = 77/239 (32%), Positives = 125/239 (52%), Gaps = 4/239 (1%)
Frame = +3
Query: 228 VRGLEEEIVCTANEIYKILEKGSAKRRTAETLLNKQSSRSHSIFSITIHIKECTPEGEEM 287
V GL E V ++++ L+ G+ R T N+ SSRSH + +T+ GE +
Sbjct: 36 VPGLIEARVYGTVDVWEKLKSGNQARSVGSTSANELSSRSHCLLRVTVL-------GENL 194
Query: 288 IKCGK----LNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINALVEHSGHVPY 343
I K L LVDLAGSE + ++ A R +E+ INKSL LG I+AL S H+PY
Sbjct: 195 INGQKTRSHLWLVDLAGSERVGKTEAEGERLKESQFINKSLSALGDVISALASKSAHIPY 374
Query: 344 RDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMK 403
R+SKLT +L+ SLGG KT + +SP L ETL +L++A R + I++ P Q +
Sbjct: 375 RNSKLTHILQSSLGGDCKTLMFVQISPGAADLTETLCSLNFATRVRGIESGPARKQTDL- 551
Query: 404 SAMIKDLYSEIDRLKQEVYAAREKNGIYIPRDRYLNEEAEKKAMAEKIERMELDADSKD 462
+E+++ KQ V ++ ++E E + + + ++ M++ +++
Sbjct: 552 --------TELNKYKQMV-------------EKVKHDEKETRKLQDNLQAMQMRLTTRE 665
>TC230674 homologue to UP|Q9M0X6 (Q9M0X6) Kinesin-like protein, partial (20%)
Length = 811
Score = 101 bits (252), Expect = 2e-21
Identities = 60/136 (44%), Positives = 81/136 (59%)
Frame = +1
Query: 250 SAKRRTAETLLNKQSSRSHSIFSITIHIKECTPEGEEMIKCGKLNLVDLAGSENISRSGA 309
S R T +N+QSSRSH F T+ I ++ ++ G LNL+DLAGSE +SRSGA
Sbjct: 85 SCSRSVGRTHMNEQSSRSH--FVXTLRISGTNSNTDQQVQ-GVLNLIDLAGSERLSRSGA 255
Query: 310 REGRAREAGEINKSLLTLGRTINALVEHSGHVPYRDSKLTRLLRDSLGGKTKTCIIATVS 369
R +E INKSL +L I AL + HVP+R+SKLT LL+ LGG +KT + +S
Sbjct: 256 TGDRLKETQAINKSLSSLSDVIFALAKKQEHVPFRNSKLTYLLQPCLGGDSKTLMFVNIS 435
Query: 370 PSIHCLEETLSTLDYA 385
P E+L +L +A
Sbjct: 436 PDPSSTGESLCSLRFA 483
>BE440889
Length = 484
Score = 100 bits (250), Expect = 3e-21
Identities = 58/134 (43%), Positives = 85/134 (63%), Gaps = 5/134 (3%)
Frame = +2
Query: 292 KLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRTINALVEHSGHVPYRDSKLTRL 351
KL LVDLAGSE ++++ + R +EA IN+SL LG I+AL S H+PYR+SKLT L
Sbjct: 86 KLWLVDLAGSERLAKTDVQGERLKEAQNINRSLSALGDVISALAAKSSHIPYRNSKLTHL 265
Query: 352 LRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKN-----IKNKPEVNQKMMKSAM 406
L+DSLGG +KT + +SPS + ETLS+L++A R + +K + + ++ AM
Sbjct: 266 LQDSLGGDSKTLMFVQISPSDQDVGETLSSLNFATRVRGVELGPVKKQIDTSEVQKMKAM 445
Query: 407 IKDLYSEIDRLKQE 420
++ SE R+K E
Sbjct: 446 LEKARSEC-RIKDE 484
>TC215161 similar to UP|Q6NQ77 (Q6NQ77) At4g05190, partial (19%)
Length = 692
Score = 98.2 bits (243), Expect = 2e-20
Identities = 56/129 (43%), Positives = 81/129 (62%), Gaps = 1/129 (0%)
Frame = +2
Query: 260 LNKQSSRSHSIFSITIH-IKECTPEGEEMIKCGKLNLVDLAGSENISRSGAREGRAREAG 318
+N+QSSRSH +F++ I+ + E T + G LNL+DLAGSE +S+SG+ R +E
Sbjct: 41 MNEQSSRSHFVFTLRIYGVNESTDXXVQ----GVLNLIDLAGSERLSKSGSTGDRLKETQ 208
Query: 319 EINKSLLTLGRTINALVEHSGHVPYRDSKLTRLLRDSLGGKTKTCIIATVSPSIHCLEET 378
INKSL +L I AL + HVP+R+SKLT LL+ LGG + T + +SP + E+
Sbjct: 209 AINKSLSSLSDVIFALAKKEDHVPFRNSKLTYLLQPCLGGYSNTLMFVNISPDPSSVGES 388
Query: 379 LSTLDYAHR 387
L +L +A R
Sbjct: 389 LCSLRFASR 415
>BE347362
Length = 448
Score = 95.5 bits (236), Expect = 1e-19
Identities = 60/152 (39%), Positives = 83/152 (54%), Gaps = 2/152 (1%)
Frame = +1
Query: 84 VQSIANKQIDRTFVFDKVFGPNSQQKELYDQAVSPIVYEVLEGYNCTIFAYGQTGTGKTY 143
+Q I + F FD VFGP Q+ ++ Q PIV VL+GYN IFAYGQTGTGKT+
Sbjct: 28 LQVICADSSKKQFKFDHVFGPEDNQETVFQQT-KPIVTSVLDGYNVCIFAYGQTGTGKTF 204
Query: 144 TMEGGAIKKNGEFPTDAGVIPRAVKQIFDILEAQ--SAEYSMKVTFLELYNEEITDLLAP 201
TMEG P GV R ++++F I E + + +Y + V+ LE+YNE+I DLL
Sbjct: 205 TMEG--------TPEHRGVNYRTLEELFRITEERHGTMKYELSVSMLEVYNEKIRDLLVE 360
Query: 202 EETTKFVDEKSKKPIALMEDGKGGVLVRGLEE 233
T + K + + + +G V GL E
Sbjct: 361 NST------QPTKKLEIKQAAEGTQEVPGLVE 438
>TC220892 homologue to UP|K125_TOBAC (O23826) 125 kDa kinesin-related
protein, partial (5%)
Length = 647
Score = 90.5 bits (223), Expect = 3e-18
Identities = 44/58 (75%), Positives = 49/58 (83%)
Frame = +3
Query: 146 EGGAIKKNGEFPTDAGVIPRAVKQIFDILEAQSAEYSMKVTFLELYNEEITDLLAPEE 203
+G + NGE P AGVIPRAVKQIFD E+Q+AEYS+KVTFLELYNEEITDLLAPEE
Sbjct: 219 KGEKLASNGELPPGAGVIPRAVKQIFDTFESQNAEYSVKVTFLELYNEEITDLLAPEE 392
>TC227748 similar to UP|Q940Y8 (Q940Y8) AT3g16060/MSL1_10, partial (34%)
Length = 1192
Score = 89.4 bits (220), Expect = 8e-18
Identities = 86/298 (28%), Positives = 140/298 (46%), Gaps = 19/298 (6%)
Frame = +3
Query: 292 KLNLVDLAGSENISRSGAREGRAR-EAGEINKSLLTLGRTINALVEHSGHVPYRDSKLTR 350
KL+ +DLAGSE + + + + R E EINKSLL L I AL GH+P+R SKLT
Sbjct: 3 KLSFIDLAGSERGADTTDNDKQTRIEGAEINKSLLALKECIRALDNDQGHIPFRGSKLTE 182
Query: 351 LLRDSLGGKTKTCIIATVSPSIHCLEETLSTLDYAHRAKNIKNKPEVNQKMMKSAM-IKD 409
+LRDS G ++T +I+ +SPS E TL+TL YA R K++ + ++ S +K+
Sbjct: 183 VLRDSFVGNSRTVMISCISPSTGSCEHTLNTLRYADRVKSLSKGNNSKKDVLSSNFNLKE 362
Query: 410 LYS-EIDRLKQEVYAAREKNG--------IYIPRDRYLNEEAEKKAMAEKIERMELDADS 460
+ + + Y R +G + P + Y + K +K +ME A +
Sbjct: 363 SSTVPLSSVTGSAYEDRVTDGWPDENEGDDFSPSEEYYE---QVKPPLKKNGKMESYATT 533
Query: 461 KDKNLVELQELYNSQQLLTAELSAKLEKTEKSLEETEQTLFDLEERHRQANAT------- 513
DK L++ S Q+ +L K+E E+ L EE A+ T
Sbjct: 534 DDK----LKK--PSGQIKWKDL-PKVEPQTTHAEDDLNALLQEEEDLVNAHRTQVEETMN 692
Query: 514 -IKEKEFLISNLLKSEKELVERAIELRAELENAASDVSNLFSKIERKDKIEEENRVLI 570
++E+ L+ + +L + L A L A+ + L +++ K +E+ VL+
Sbjct: 693 IVREEMNLLVEADQPGNQLDDYITRLNAILSQKAAGILQLQTRLAHFQKRLKEHNVLV 866
>BE802280 similar to GP|15186760|gb KRP120-2 {Daucus carota}, partial (2%)
Length = 311
Score = 86.3 bits (212), Expect = 7e-17
Identities = 41/51 (80%), Positives = 45/51 (87%)
Frame = +3
Query: 999 EELRTPSFEELLKAFWDAKSQKLANGDVKHIGSYEETQSVRDSRVPLTTIN 1049
EELRTPSFEELLK+FWDAKS K ANGDV++IG+YE QSVRDSRVPL IN
Sbjct: 3 EELRTPSFEELLKSFWDAKSPKHANGDVRYIGAYEAAQSVRDSRVPLVAIN 155
>BU964987 similar to GP|15208447|gb kinesin heavy chain {Zea mays}, partial
(19%)
Length = 443
Score = 82.4 bits (202), Expect = 9e-16
Identities = 55/153 (35%), Positives = 80/153 (51%), Gaps = 2/153 (1%)
Frame = +2
Query: 184 KVTFLELYNEEITDLLAPEETTKFVDEKSKKPIALMEDGKGGVLVRGLEEEIVCTANEIY 243
K + +E+YNE I DLL+ E T+ + L +D + G +V L EE + +
Sbjct: 2 KFSAIEIYNEIIRDLLSTENTS----------LRLRDDPERGPIVEKLTEETLRNWVHLK 151
Query: 244 KILEKGSAKRRTAETLLNKQSSRSHSIFSITIH--IKECTPEGEEMIKCGKLNLVDLAGS 301
++L A+R+ ET LN +SSRSH I +TI +E + +N VDLAGS
Sbjct: 152 ELLSFCEAQRQVGETYLNDKSSRSHQIIRLTIESSAREFMGKSSSTTLAASVNFVDLAGS 331
Query: 302 ENISRSGAREGRAREAGEINKSLLTLGRTINAL 334
E S++ + R +E IN+SLLTLG I L
Sbjct: 332 ERASQALSAGARLKEGCHINRSLLTLGTVIRKL 430
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.311 0.127 0.341
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 34,284,218
Number of Sequences: 63676
Number of extensions: 388444
Number of successful extensions: 1616
Number of sequences better than 10.0: 93
Number of HSP's better than 10.0 without gapping: 1574
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1589
length of query: 1049
length of database: 12,639,632
effective HSP length: 107
effective length of query: 942
effective length of database: 5,826,300
effective search space: 5488374600
effective search space used: 5488374600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC149490.9


