

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
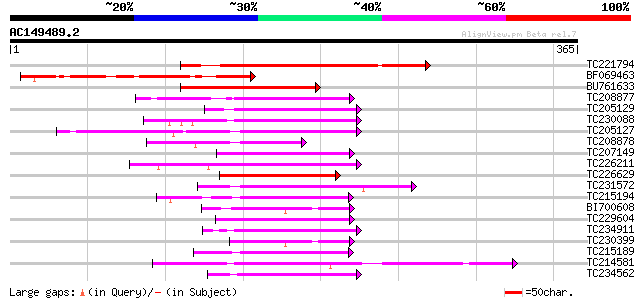
Query= AC149489.2 - phase: 0
(365 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC221794 homologue to UP|Q9FR59 (Q9FR59) Homeobox 1, partial (20%) 164 6e-41
BF069463 similar to GP|13346176|gb| BNLGHi8377 {Gossypium hirsut... 112 3e-25
BU761633 similar to GP|1814424|gb|A homeodomain protein AHDP {Ar... 101 6e-22
TC208877 similar to UP|Q39862 (Q39862) Homeobox-leucine zipper p... 78 5e-15
TC205129 homologue to UP|HT22_ARATH (P46604) Homeobox-leucine zi... 69 4e-12
TC230088 UP|Q9SP47 (Q9SP47) Homeodomain-leucine zipper protein 5... 67 1e-11
TC205127 similar to UP|HT22_ARATH (P46604) Homeobox-leucine zipp... 67 1e-11
TC208878 similar to UP|Q39862 (Q39862) Homeobox-leucine zipper p... 67 2e-11
TC207149 similar to UP|Q9XH73 (Q9XH73) Homeobox leucine zipper p... 66 2e-11
TC226211 UP|Q39862 (Q39862) Homeobox-leucine zipper protein, com... 65 4e-11
TC226629 homologue to UP|Q93XA3 (Q93XA3) Homeodomain leucine zip... 65 4e-11
TC231572 similar to UP|HT14_ARATH (P46665) Homeobox-leucine zipp... 65 6e-11
TC215194 similar to UP|Q39941 (Q39941) HAHB-1, partial (56%) 64 8e-11
BI700608 homologue to GP|19578319|em homeodomain-leucine zipper ... 64 1e-10
TC229604 similar to UP|Q9M276 (Q9M276) Homeobox-leucine zipper p... 64 1e-10
TC234911 similar to UP|HT22_ARATH (P46604) Homeobox-leucine zipp... 64 1e-10
TC230399 homologue to UP|Q6JE95 (Q6JE95) Class III HD-Zip protei... 62 4e-10
TC215189 similar to UP|Q39941 (Q39941) HAHB-1, partial (70%) 60 2e-09
TC214581 similar to UP|Q43427 (Q43427) DNA-binding protein, part... 60 2e-09
TC234562 similar to UP|HT22_ARATH (P46604) Homeobox-leucine zipp... 59 3e-09
>TC221794 homologue to UP|Q9FR59 (Q9FR59) Homeobox 1, partial (20%)
Length = 501
Score = 164 bits (415), Expect = 6e-41
Identities = 81/161 (50%), Positives = 106/161 (65%)
Frame = +3
Query: 111 DELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQ 170
+ LE GED + + K+K+YHRHT QI+ MEA FKE PHPD+KQR+
Sbjct: 21 ENLEGASGEDQDP------------RPNKKKRYHRHTQHQIQEMEAFFKECPHPDDKQRK 164
Query: 171 QLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINKACCP 230
+LS++LGL P QVKFWFQN+RTQ+K ERHEN+ L++E EKLR N RE + A CP
Sbjct: 165 ELSRELGLEPLQVKFWFQNKRTQMKTQHERHENTQLRTENEKLRADNMRFREALGNASCP 344
Query: 231 NCGVPTTNRDGTMATEEQQLRIENAKLKAEVERLRAALGKY 271
NCG PT G M+ +E LR+ENA+L+ E++R+ A KY
Sbjct: 345 NCGGPTA--IGEMSFDEHHLRLENARLREEIDRISAIAAKY 461
>BF069463 similar to GP|13346176|gb| BNLGHi8377 {Gossypium hirsutum}, partial
(10%)
Length = 419
Score = 112 bits (279), Expect = 3e-25
Identities = 75/154 (48%), Positives = 94/154 (60%), Gaps = 3/154 (1%)
Frame = +1
Query: 8 LLKKHNT---LLSSPTLSMCADMSNNNPPSTSKAKDFFPSPALSLSLAGIFRHGGGAAAE 64
LLKKH+ LSM +MSN PP T KD F S +LSLSLAGIFRH G AA
Sbjct: 28 LLKKHHQEQQKAQQTWLSMGVNMSNT-PPHT---KDLFSSSSLSLSLAGIFRHAGEAA-- 189
Query: 65 GEGGSISNMEVEEGEEGSTIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGD 124
S MEV G E + E EISS+ SGP +S+S ++++G+ L +D DD+GD
Sbjct: 190 ----ETSGMEVAGGNE---VRREDTVEISSDNSGPVRSRSEEDFDGEGLHED---DDDGD 339
Query: 125 GDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALF 158
+KN+KKK++KYHRHT+EQIR MEALF
Sbjct: 340 --------DKNRKKKKRKYHRHTAEQIREMEALF 417
>BU761633 similar to GP|1814424|gb|A homeodomain protein AHDP {Arabidopsis
thaliana}, partial (12%)
Length = 379
Score = 101 bits (251), Expect = 6e-22
Identities = 48/90 (53%), Positives = 60/90 (66%)
Frame = +1
Query: 111 DELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQ 170
DE E G D+ GD ++ +++K+YHRHT QI+ +EA FKE PHPDEKQR
Sbjct: 109 DEYESRSGSDNFEGASGDDQDGGDDQPQRKKRYHRHTPHQIQELEAFFKECPHPDEKQRL 288
Query: 171 QLSKQLGLAPRQVKFWFQNRRTQIKAIQER 200
LSK+L L +QVKFWFQNRRTQ+K ER
Sbjct: 289 DLSKRLALENKQVKFWFQNRRTQMKTQLER 378
>TC208877 similar to UP|Q39862 (Q39862) Homeobox-leucine zipper protein,
partial (46%)
Length = 768
Score = 78.2 bits (191), Expect = 5e-15
Identities = 54/141 (38%), Positives = 72/141 (50%)
Frame = +3
Query: 82 STIGGERVEEISSEYSGPAKSKSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRK 141
S+I G+R E E +G ++ G +DDEG GD D D RK
Sbjct: 381 SSISGKRSER---EGNGEENERTSSSRGGGGSDDDEGGACGGDADADAS---------RK 524
Query: 142 KYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERH 201
K R + EQ V+E FKE + KQ+Q L+KQL L PRQV+ WFQNRR + K Q
Sbjct: 525 KL-RLSKEQALVLEETFKEHNTLNPKQKQALAKQLNLMPRQVEVWFQNRRARTKLKQTEV 701
Query: 202 ENSLLKSEIEKLREKNKTLRE 222
+ LK E L E+N+ L++
Sbjct: 702 DCEYLKRCCENLTEENRRLQK 764
>TC205129 homologue to UP|HT22_ARATH (P46604) Homeobox-leucine zipper protein
HAT22 (HD-ZIP protein 22), partial (47%)
Length = 793
Score = 68.6 bits (166), Expect = 4e-12
Identities = 39/101 (38%), Positives = 56/101 (54%)
Frame = +3
Query: 126 DGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKF 185
D D DG N KK R T EQ ++E FK+ + KQ+Q L++QL L PRQV+
Sbjct: 105 DEDEDGTNARKKL------RLTKEQSALLEESFKQHSTLNPKQKQALARQLNLRPRQVEV 266
Query: 186 WFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINK 226
WFQNRR + K Q + LK E L ++N+ L++ + +
Sbjct: 267 WFQNRRARTKLKQTEVDCEFLKKCCETLTDENRRLKKELQE 389
>TC230088 UP|Q9SP47 (Q9SP47) Homeodomain-leucine zipper protein 57, complete
Length = 1105
Score = 67.4 bits (163), Expect = 1e-11
Identities = 47/149 (31%), Positives = 80/149 (53%), Gaps = 9/149 (6%)
Frame = +1
Query: 87 ERVEEISSEYSGPAK---SKSIDEYE---GDELEDD---EGEDDEGDGDGDGDGVNKNKK 137
E +E + ++ S PA SK + ++E G + D + + E + D D +G
Sbjct: 49 EVLESLWAQTSNPASFQGSKPVVDFENVSGSRMTDRPFFQALEKEENCDEDYEGCFHQPG 228
Query: 138 KKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAI 197
KKR R TSEQ++ +E F+ + +++ QL+K+LGL PRQV WFQNRR + K
Sbjct: 229 KKR----RLTSEQVQFLERNFEVENKLEPERKVQLAKELGLQPRQVAIWFQNRRARFKTK 396
Query: 198 QERHENSLLKSEIEKLREKNKTLRETINK 226
Q + +LK+ ++L+ ++L + +K
Sbjct: 397 QLEKDYGVLKASYDRLKSDYESLVQENDK 483
>TC205127 similar to UP|HT22_ARATH (P46604) Homeobox-leucine zipper protein
HAT22 (HD-ZIP protein 22), partial (59%)
Length = 1036
Score = 67.0 bits (162), Expect = 1e-11
Identities = 57/203 (28%), Positives = 89/203 (43%), Gaps = 7/203 (3%)
Frame = +3
Query: 31 NPPSTSKAKDFFPSPALSLSLAGIFRHGGGAAAEGEGGSISNMEVEEGEEGSTIGGERVE 90
+P ++S+A+ PS L LS + + +S+ + +
Sbjct: 18 SPYNSSEAE---PSLTLGLSRESYLKVPKNIIGQNSNNKVSSCDDPLDHLSTQTNSPHHS 188
Query: 91 EISSEYSGPAKSKS-------IDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKY 143
+SS SG K + +D E D+ D E D D DG KK + K
Sbjct: 189 AVSSFSSGRVKRERDLSCEEVVDATEIDQ-RDHSCEGIVRATDEDEDGTAARKKLRLSK- 362
Query: 144 HRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHEN 203
EQ ++E FK+ + KQ+Q L+KQL L PRQV+ WFQNRR + K Q +
Sbjct: 363 -----EQSALLEESFKQHSTLNPKQKQALAKQLNLRPRQVEVWFQNRRARTKLKQTEVDC 527
Query: 204 SLLKSEIEKLREKNKTLRETINK 226
LK E L ++N+ L++ + +
Sbjct: 528 EFLKKCCETLTDENRRLQKELQE 596
>TC208878 similar to UP|Q39862 (Q39862) Homeobox-leucine zipper protein,
partial (46%)
Length = 701
Score = 66.6 bits (161), Expect = 2e-11
Identities = 45/110 (40%), Positives = 55/110 (49%), Gaps = 7/110 (6%)
Frame = +1
Query: 89 VEEISSEYSGPAKSKSIDEYEGDELEDDEG-------EDDEGDGDGDGDGVNKNKKKKRK 141
V +S S + +S E GDE E +DD G GDGDG KK +
Sbjct: 274 VSSPNSTISSISGKRSEREGNGDENERTSSSRGGGSDDDDGGACGGDGDGEASRKKLRLS 453
Query: 142 KYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRR 191
K EQ V+E FKE + KQ+Q L+KQL L PRQV+ WFQNRR
Sbjct: 454 K------EQALVLEETFKEHNSLNPKQKQALAKQLNLMPRQVEVWFQNRR 585
>TC207149 similar to UP|Q9XH73 (Q9XH73) Homeobox leucine zipper protein,
partial (42%)
Length = 873
Score = 66.2 bits (160), Expect = 2e-11
Identities = 32/89 (35%), Positives = 53/89 (58%)
Frame = +2
Query: 134 KNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQ 193
+ K KK + R + EQIR +E +F+ + +++ QL++ LGL PRQV WFQNRR +
Sbjct: 437 RKKSKKIENKRRFSDEQIRSLECIFESESKLEPRKKMQLARDLGLQPRQVAIWFQNRRAR 616
Query: 194 IKAIQERHENSLLKSEIEKLREKNKTLRE 222
K+ + E LK E + L + ++L++
Sbjct: 617 WKSKRIEQEYRKLKDEYDNLASRFESLKK 703
>TC226211 UP|Q39862 (Q39862) Homeobox-leucine zipper protein, complete
Length = 1249
Score = 65.5 bits (158), Expect = 4e-11
Identities = 49/157 (31%), Positives = 75/157 (47%), Gaps = 8/157 (5%)
Frame = +3
Query: 78 GEEGSTIGGERVEEISS-----EYSGPAKSKSIDEYEGDELEDDEGEDDEGDGD---GDG 129
GE GS + G V + S E +G + S + + E +E D D G
Sbjct: 261 GEAGSFLRGIDVNRLPSVVDCEEEAGVSSPNSTVSSVSGKRSERETNGEENDTDRACSRG 440
Query: 130 DGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQN 189
++ + +K R + +Q V+E FKE + KQ+ L+KQLGL RQV+ WFQN
Sbjct: 441 IISDEEDAETSRKKLRLSKDQSIVLEESFKEHNTLNPKQKLALAKQLGLRARQVEVWFQN 620
Query: 190 RRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINK 226
RR + K Q + LK E L E+N+ L++ + +
Sbjct: 621 RRARTKLKQTEVDCEFLKRCCENLTEENRRLQKEVQE 731
>TC226629 homologue to UP|Q93XA3 (Q93XA3) Homeodomain leucine zipper protein
HDZ3 (Fragment), partial (96%)
Length = 975
Score = 65.5 bits (158), Expect = 4e-11
Identities = 32/78 (41%), Positives = 50/78 (64%)
Frame = +3
Query: 136 KKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIK 195
+K+ +K HR +SEQ+ ++E F+E + +++ QL+K+LGL PRQV WFQNRR + K
Sbjct: 69 EKQSPEKKHRLSSEQVHLLEKSFEEENKLEPERKTQLAKKLGLQPRQVAVWFQNRRARWK 248
Query: 196 AIQERHENSLLKSEIEKL 213
Q + +LKS + L
Sbjct: 249 TKQLERDYDVLKSSYDTL 302
>TC231572 similar to UP|HT14_ARATH (P46665) Homeobox-leucine zipper protein
HAT14 (HD-ZIP protein 14), partial (53%)
Length = 835
Score = 64.7 bits (156), Expect = 6e-11
Identities = 47/160 (29%), Positives = 68/160 (42%), Gaps = 19/160 (11%)
Frame = +3
Query: 122 EGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPR 181
+ D + +G G N KK + K EQ +E FKE + KQ+ L+KQL L PR
Sbjct: 24 DDDDNNNGSGGNTRKKLRLSK------EQSAFLEESFKEHNTLNPKQKLALAKQLNLQPR 185
Query: 182 QVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINK--------------- 226
QV+ WF NRR + K Q + LK E L E+N+ L + + +
Sbjct: 186 QVEVWFFNRRARTKLKQTEVDCEYLKRCCETLTEENRRLHKELQELRALKTSNPFYMQLP 365
Query: 227 ----ACCPNCGVPTTNRDGTMATEEQQLRIENAKLKAEVE 262
CP+C TN T + + N+ A V+
Sbjct: 366 ATTLTMCPSCERVATNSTSTSLSISATINATNSGAMASVK 485
>TC215194 similar to UP|Q39941 (Q39941) HAHB-1, partial (56%)
Length = 1060
Score = 64.3 bits (155), Expect = 8e-11
Identities = 40/130 (30%), Positives = 65/130 (49%), Gaps = 3/130 (2%)
Frame = +2
Query: 95 EYSGPAKS---KSIDEYEGDELEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQI 151
EY G S K + G EL ++ +++ D DG +KK+R EQ+
Sbjct: 101 EYHGGGASFLGKRSMSFSGIELGEEANAEEDSD-----DGSQAGEKKRRLNM-----EQV 250
Query: 152 RVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKSEIE 211
+ +E F+ + +++ QL++ LGL PRQ+ WFQNRR + K Q + LLK + E
Sbjct: 251 KTLEKSFELGNKLEPERKMQLARALGLQPRQIAIWFQNRRARWKTKQLEKDYDLLKRQYE 430
Query: 212 KLREKNKTLR 221
++ N L+
Sbjct: 431 AIKADNDALQ 460
>BI700608 homologue to GP|19578319|em homeodomain-leucine zipper protein
{Arabidopsis thaliana}, partial (13%)
Length = 421
Score = 63.9 bits (154), Expect = 1e-10
Identities = 38/103 (36%), Positives = 57/103 (54%), Gaps = 4/103 (3%)
Frame = +3
Query: 124 DGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQL----GLA 179
DG +G G++ K Y R+T EQ+ +E L+ + P P +RQQL ++ +
Sbjct: 60 DGSRNGIGMDNGK------YVRYTPEQVEALERLYHDCPKPSSIRRQQLIRECPILSNIE 221
Query: 180 PRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRE 222
P+Q+K WFQNRR + K +R E+S L++ KL NK L E
Sbjct: 222 PKQIKVWFQNRRCREK---QRKESSRLQAVNRKLTAMNKLLME 341
>TC229604 similar to UP|Q9M276 (Q9M276) Homeobox-leucine zipper protein
ATHB-12, partial (44%)
Length = 992
Score = 63.5 bits (153), Expect = 1e-10
Identities = 28/90 (31%), Positives = 54/90 (59%)
Frame = +2
Query: 133 NKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRT 192
+++KK+ R + EQI+ +E +F+ + +++ QL+++LGL PRQV WFQN+R
Sbjct: 206 SRSKKRNNNNTRRFSDEQIKSLETMFESETRLEPRKKLQLARELGLQPRQVAIWFQNKRA 385
Query: 193 QIKAIQERHENSLLKSEIEKLREKNKTLRE 222
+ K+ Q + +L+S L + + L++
Sbjct: 386 RWKSKQLERDYGILQSNYNTLASRFEALKK 475
>TC234911 similar to UP|HT22_ARATH (P46604) Homeobox-leucine zipper protein
HAT22 (HD-ZIP protein 22), partial (40%)
Length = 799
Score = 63.5 bits (153), Expect = 1e-10
Identities = 40/102 (39%), Positives = 57/102 (55%)
Frame = +1
Query: 125 GDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVK 184
GD D DG N +KK R T EQ V+E F+E + KQ+Q+L+ +L L RQV+
Sbjct: 85 GDVDEDG---NPRKKL----RLTKEQAAVLEENFREHSTLNPKQKQELAMKLNLRARQVE 243
Query: 185 FWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINK 226
WFQNRR + K Q + LLK + L +NK L++ + +
Sbjct: 244 VWFQNRRARTKLKQTESDCELLKKCCDTLTVENKKLQKELQE 369
>TC230399 homologue to UP|Q6JE95 (Q6JE95) Class III HD-Zip protein, partial
(27%)
Length = 1334
Score = 62.0 bits (149), Expect = 4e-10
Identities = 34/85 (40%), Positives = 50/85 (58%), Gaps = 4/85 (4%)
Frame = +3
Query: 142 KYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQL----GLAPRQVKFWFQNRRTQIKAI 197
KY R+T+EQ+ +E ++ E P P +RQQL ++ + P+Q+K WFQNRR + K
Sbjct: 699 KYVRYTAEQVEALERVYAECPKPSSLRRQQLIRECPILSNIEPKQIKVWFQNRRCREK-- 872
Query: 198 QERHENSLLKSEIEKLREKNKTLRE 222
+R E S L++ KL NK L E
Sbjct: 873 -QRKEASRLQTVNRKLTAMNKLLME 944
>TC215189 similar to UP|Q39941 (Q39941) HAHB-1, partial (70%)
Length = 1315
Score = 59.7 bits (143), Expect = 2e-09
Identities = 32/103 (31%), Positives = 54/103 (52%)
Frame = +3
Query: 119 EDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGL 178
E + + D DG +KK+R EQ++ +E F+ + +++ QL++ LGL
Sbjct: 408 EANNAEEDLSDDGSQAGEKKRRLNM-----EQVKTLEKSFELGNKLEPERKMQLARALGL 572
Query: 179 APRQVKFWFQNRRTQIKAIQERHENSLLKSEIEKLREKNKTLR 221
PRQ+ WFQNRR + K Q + +LK + E ++ N L+
Sbjct: 573 QPRQIAIWFQNRRARWKTKQLEKDYDVLKRQYEAVKSDNDALQ 701
>TC214581 similar to UP|Q43427 (Q43427) DNA-binding protein, partial (54%)
Length = 1605
Score = 59.7 bits (143), Expect = 2e-09
Identities = 71/246 (28%), Positives = 110/246 (43%), Gaps = 11/246 (4%)
Frame = +2
Query: 93 SSEYSGPAKSKSIDEYEGDE-LEDDEGEDDEGDGDGDGDGVNKNKKKKRKKYHRHTSEQI 151
SS + G S + +G + D + +GD D D +KKR R + EQ+
Sbjct: 443 SSPFLGSRSIVSFGDVQGGKGCNDSFFRPYDENGDEDMDEYFHQPEKKR----RLSVEQV 610
Query: 152 RVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQ-ERHENSL----- 205
+ +E F E + +++ L+K+LGL PRQV WFQNRR + K Q E+ +SL
Sbjct: 611 KFLEKSFDEENKLEPERKIWLAKELGLQPRQVAIWFQNRRARWKTKQMEKDYDSLQASYN 790
Query: 206 -LKSEIEK-LREKNKTLRETINKACCPNCGVPTTNRDGTMATEEQQLRIENAKLKAEVER 263
LK+ + LREK+K E + + E+ + +E A+ E
Sbjct: 791 DLKANYDNLLREKDKLKAEVARLT------------EKVLGREKNESHLEQAETNGLQEP 934
Query: 264 LRAALGKYAS-GTMSPSCSTSHDQENIKSSLDFYTGIFCLDES-RIMDVVNQAMEELIKM 321
L +L AS G S + QE+I S+ + IF ES + D V+ A+ E
Sbjct: 935 LHKSLVDSASEGEGSKVTFEACKQEDISSA---KSDIFDSSESPQYTDGVHSALLETGDS 1105
Query: 322 ATMGEP 327
+ + EP
Sbjct: 1106SYVFEP 1123
>TC234562 similar to UP|HT22_ARATH (P46604) Homeobox-leucine zipper protein
HAT22 (HD-ZIP protein 22), partial (33%)
Length = 585
Score = 58.9 bits (141), Expect = 3e-09
Identities = 35/99 (35%), Positives = 52/99 (52%)
Frame = +3
Query: 128 DGDGVNKNKKKKRKKYHRHTSEQIRVMEALFKESPHPDEKQRQQLSKQLGLAPRQVKFWF 187
D + N +KK K T EQ +E +FK + Q+Q L++QL L RQV+ WF
Sbjct: 270 DSNNSNNGSRKKLKL----TKEQSATLEDIFKLHSSLNPAQKQALAEQLNLKHRQVEVWF 437
Query: 188 QNRRTQIKAIQERHENSLLKSEIEKLREKNKTLRETINK 226
QNRR + K Q + LK EKL ++N L++ + +
Sbjct: 438 QNRRARTKLKQTEVDCEFLKKCCEKLTDENLRLKKELQE 554
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.309 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,182,634
Number of Sequences: 63676
Number of extensions: 206563
Number of successful extensions: 2950
Number of sequences better than 10.0: 352
Number of HSP's better than 10.0 without gapping: 2157
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2575
length of query: 365
length of database: 12,639,632
effective HSP length: 99
effective length of query: 266
effective length of database: 6,335,708
effective search space: 1685298328
effective search space used: 1685298328
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC149489.2


