

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149305.9 - phase: 0 /pseudo
(136 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
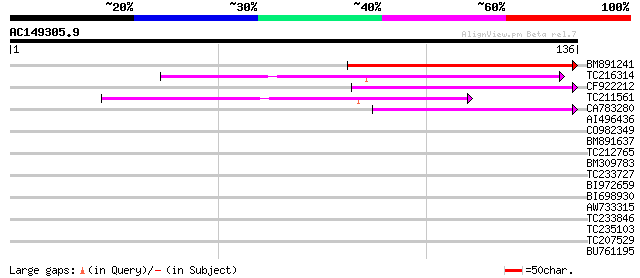
Score E
Sequences producing significant alignments: (bits) Value
BM891241 49 1e-06
TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {A... 44 3e-05
CF922212 43 5e-05
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 42 7e-05
CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polym... 39 6e-04
AI496436 36 0.005
CO982349 34 0.024
BM891637 33 0.042
TC212765 31 0.16
BM309783 31 0.21
TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR ret... 30 0.27
BI972659 30 0.35
BI698930 29 0.78
AW733315 26 5.1
TC233846 26 5.1
TC235103 similar to UP|Q6NKU9 (Q6NKU9) At5g55510, partial (9%) 26 6.6
TC207529 similar to UP|P93045 (P93045) SNF5 homolog BSH, partial... 25 8.7
BU761195 25 8.7
>BM891241
Length = 407
Score = 48.5 bits (114), Expect = 1e-06
Identities = 21/55 (38%), Positives = 34/55 (61%)
Frame = -2
Query: 82 NKGMNMVGWDKITKAKKNGGLGVRVVRKQNIALLGKLVWNLLMPTNHLWSQILSA 136
+K + V W+ + K GGLG++ + K NIALLGK +W L LW++I+++
Sbjct: 391 HKKIPWVKWEVVCLPKTKGGLGIKDISKFNIALLGKWIWALASDQQQLWARIINS 227
>TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {Arabidopsis
thaliana;} , partial (18%)
Length = 759
Score = 43.5 bits (101), Expect = 3e-05
Identities = 28/98 (28%), Positives = 48/98 (48%), Gaps = 1/98 (1%)
Frame = +2
Query: 37 RVILVNSVLTSIPAYKMQIH*LAHYICVIDHLNKTVHSFI*K*STNKG-MNMVGWDKITK 95
RV L+ SVL+++P + + I +D L F+ + + + V W I
Sbjct: 11 RVTLIKSVLSALPIXXLSFFKIPQRI--VDKLVTLQRQFLWGGTQHHNRIPWVKWADICN 184
Query: 96 AKKNGGLGVRVVRKQNIALLGKLVWNLLMPTNHLWSQI 133
K +GGLG++ + N AL G+ +W L N LW+++
Sbjct: 185 PKIDGGLGIKDLSNFNAALRGRWIWGLASNHNQLWARL 298
>CF922212
Length = 445
Score = 42.7 bits (99), Expect = 5e-05
Identities = 22/54 (40%), Positives = 30/54 (54%)
Frame = -2
Query: 83 KGMNMVGWDKITKAKKNGGLGVRVVRKQNIALLGKLVWNLLMPTNHLWSQILSA 136
K + V WD + K+ GLG R +RK N ALLGK WNL L +++L +
Sbjct: 369 KKIAWVNWDSVCLPKEKEGLGNRDLRKFNYALLGKRRWNLFHHQGELGARVLDS 208
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase
(Fragment) , partial (59%)
Length = 1077
Score = 42.4 bits (98), Expect = 7e-05
Identities = 29/90 (32%), Positives = 46/90 (50%), Gaps = 1/90 (1%)
Frame = +1
Query: 23 KLASWKCRLLNKPSRVILVNSVLTSIPAYKMQIH*LAHYICVIDHLNKTVHSFI*K*STN 82
KL+ WK R ++ RV L+ SVLTSIP Y + V+D L K +F+ +
Sbjct: 673 KLSKWK*RHISFGGRVTLIKSVLTSIPIYFFSFFRVPQ--SVVDKLVKI*RTFLWGGGPD 846
Query: 83 -KGMNMVGWDKITKAKKNGGLGVRVVRKQN 111
K ++ + W+K+ K+ GG+ R+ N
Sbjct: 847 QKKISWIRWEKLCLPKERGGI*KRLPNNHN 936
>CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polymerase
homolog T24M8.8 - Arabidopsis thaliana, partial (4%)
Length = 456
Score = 39.3 bits (90), Expect = 6e-04
Identities = 17/49 (34%), Positives = 26/49 (52%)
Frame = +2
Query: 88 VGWDKITKAKKNGGLGVRVVRKQNIALLGKLVWNLLMPTNHLWSQILSA 136
V W + K GGLG++ +R N LLGK W+L W+++L +
Sbjct: 104 VNWKTVCLPKAKGGLGIKDLRTFNTTLLGKWRWDLFYIQQEPWAKVLQS 250
>AI496436
Length = 414
Score = 36.2 bits (82), Expect = 0.005
Identities = 16/29 (55%), Positives = 22/29 (75%)
Frame = +1
Query: 23 KLASWKCRLLNKPSRVILVNSVLTSIPAY 51
KLASWK + ++ RV L+NSVLT++P Y
Sbjct: 220 KLASWKQKYVSFGGRVTLINSVLTALPIY 306
>CO982349
Length = 795
Score = 33.9 bits (76), Expect = 0.024
Identities = 14/29 (48%), Positives = 21/29 (72%)
Frame = +3
Query: 23 KLASWKCRLLNKPSRVILVNSVLTSIPAY 51
KLA WK R ++ RV L+N++LT++P Y
Sbjct: 576 KLARWKQRHISLGGRVTLINAILTALPIY 662
>BM891637
Length = 427
Score = 33.1 bits (74), Expect = 0.042
Identities = 16/52 (30%), Positives = 28/52 (53%)
Frame = -1
Query: 83 KGMNMVGWDKITKAKKNGGLGVRVVRKQNIALLGKLVWNLLMPTNHLWSQIL 134
K + + W + + GGLG++ ++ N ALL K W + + LW++IL
Sbjct: 418 KKIAWISWRQCCASGDVGGLGIKDIKILNNALLIKWKWLMFHQPHQLWNRIL 263
>TC212765
Length = 637
Score = 31.2 bits (69), Expect = 0.16
Identities = 17/38 (44%), Positives = 22/38 (57%)
Frame = +2
Query: 17 CGFLINKLASWKCRLLNKPSRVILVNSVLTSIPAYKMQ 54
CG LASWK +LLN+ S+ L VL + P Y M+
Sbjct: 395 CGAHP*PLASWKGKLLNQASKHCLT*LVLHAFPIYPME 508
>BM309783
Length = 433
Score = 30.8 bits (68), Expect = 0.21
Identities = 15/49 (30%), Positives = 25/49 (50%)
Frame = +2
Query: 86 NMVGWDKITKAKKNGGLGVRVVRKQNIALLGKLVWNLLMPTNHLWSQIL 134
++VGW + +GG+G+ + N A L K+ W M LW+ +L
Sbjct: 35 HVVGWKAMIMP*HSGGVGMCDLTIMNQACLMKIGWAFRMGDGSLWADVL 181
>TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR retroelement
reverse transcriptase, partial (7%)
Length = 956
Score = 30.4 bits (67), Expect = 0.27
Identities = 14/34 (41%), Positives = 21/34 (61%)
Frame = -2
Query: 23 KLASWKCRLLNKPSRVILVNSVLTSIPAYKMQIH 56
KLASWK +L+ R+ LVNSV+ + Y ++
Sbjct: 160 KLASWKGSILSIMGRIQLVNSVIHGMLLYSFSVY 59
>BI972659
Length = 453
Score = 30.0 bits (66), Expect = 0.35
Identities = 19/68 (27%), Positives = 32/68 (46%)
Frame = +1
Query: 23 KLASWKCRLLNKPSRVILVNSVLTSIPAYKMQIH*LAHYICVIDHLNKTVHSFI*K*STN 82
KL+ W + L+ RV L+ SVL ++P Y + + I +D L +F+ N
Sbjct: 259 KLSKWNQKYLSMGGRVTLIKSVLNALPIYLLSFFKIPQRI--VDKLVSLQRTFM--WGGN 426
Query: 83 KGMNMVGW 90
+ N + W
Sbjct: 427 QHHNRISW 450
>BI698930
Length = 384
Score = 28.9 bits (63), Expect = 0.78
Identities = 12/28 (42%), Positives = 19/28 (67%)
Frame = +3
Query: 24 LASWKCRLLNKPSRVILVNSVLTSIPAY 51
LA WK +L+ R L+NS+L+S+P +
Sbjct: 81 LALWKHEVLSFAGRTCLINSMLSSLPLF 164
>AW733315
Length = 333
Score = 26.2 bits (56), Expect = 5.1
Identities = 10/19 (52%), Positives = 13/19 (67%)
Frame = +2
Query: 9 IFKLISLVCGFLINKLASW 27
+FK+I L C L+ KL SW
Sbjct: 257 LFKIILLYCSILLPKLLSW 313
>TC233846
Length = 424
Score = 26.2 bits (56), Expect = 5.1
Identities = 10/20 (50%), Positives = 15/20 (75%)
Frame = -1
Query: 112 IALLGKLVWNLLMPTNHLWS 131
I+LLGKLVW L+ + +W+
Sbjct: 289 ISLLGKLVWTLVSGCSTVWA 230
>TC235103 similar to UP|Q6NKU9 (Q6NKU9) At5g55510, partial (9%)
Length = 872
Score = 25.8 bits (55), Expect = 6.6
Identities = 12/31 (38%), Positives = 20/31 (63%)
Frame = -2
Query: 4 NHFNIIFKLISLVCGFLINKLASWKCRLLNK 34
N+ N+ K I + FL+NKL +++ RL+ K
Sbjct: 556 NNKNVQEKKIKYIFFFLVNKLHTYRDRLIQK 464
>TC207529 similar to UP|P93045 (P93045) SNF5 homolog BSH, partial (82%)
Length = 1265
Score = 25.4 bits (54), Expect = 8.7
Identities = 13/41 (31%), Positives = 20/41 (48%)
Frame = +3
Query: 23 KLASWKCRLLNKPSRVILVNSVLTSIPAYKMQIH*LAHYIC 63
+L C L PSR+ N + SI A+ + + * Y+C
Sbjct: 237 RLCKKDC*RLEAPSRLCNTNCSINSITAFGVSVL*RPGYVC 359
>BU761195
Length = 472
Score = 25.4 bits (54), Expect = 8.7
Identities = 9/21 (42%), Positives = 14/21 (65%)
Frame = -3
Query: 56 H*LAHYICVIDHLNKTVHSFI 76
H + YIC++ H+ +HSFI
Sbjct: 143 HHIYIYICILPHIYIPIHSFI 81
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.337 0.146 0.476
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,283,781
Number of Sequences: 63676
Number of extensions: 93864
Number of successful extensions: 865
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 862
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 864
length of query: 136
length of database: 12,639,632
effective HSP length: 87
effective length of query: 49
effective length of database: 7,099,820
effective search space: 347891180
effective search space used: 347891180
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 54 (25.4 bits)
Medicago: description of AC149305.9


