

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149207.4 - phase: 0
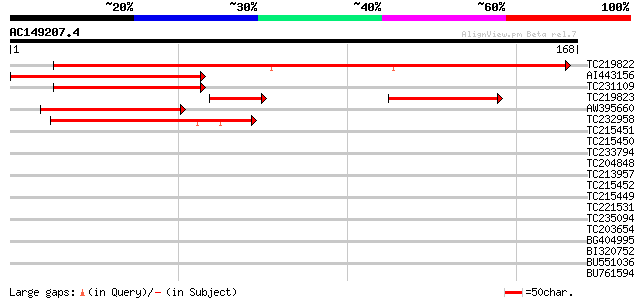
(168 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC219822 homologue to UP|CENB_HUMAN (P07199) Major centromere au... 164 2e-41
AI443156 52 1e-07
TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, par... 52 2e-07
TC219823 weakly similar to UP|Q7RJ53 (Q7RJ53) Arabidopsis thalia... 45 2e-06
AW395660 44 5e-05
TC232958 41 2e-04
TC215451 weakly similar to UP|Q94K07 (Q94K07) Nucleosome assembl... 39 0.001
TC215450 similar to UP|P93488 (P93488) NAP1Ps, partial (30%) 39 0.001
TC233794 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {A... 38 0.002
TC204848 similar to UP|Q6ICZ8 (Q6ICZ8) At5g13850, partial (89%) 38 0.003
TC213957 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {A... 38 0.003
TC215452 similar to UP|P93488 (P93488) NAP1Ps, partial (29%) 38 0.003
TC215449 similar to UP|P93488 (P93488) NAP1Ps, partial (97%) 37 0.004
TC221531 similar to UP|Q84N48 (Q84N48) CRS2-associated factor 2,... 37 0.006
TC235094 similar to UP|PRT3_SCYCA (P30258) Protamine Z3 (Scyllio... 37 0.006
TC203654 similar to PIR|T46225|T46225 alpha NAC-like protein - A... 37 0.006
BG404995 similar to GP|22831068|dbj OJ1131_E05.32 {Oryza sativa ... 36 0.008
BI320752 35 0.013
BU551036 35 0.013
BU761594 similar to GP|22654971|gb At1g80930/F23A5_23 {Arabidops... 35 0.013
>TC219822 homologue to UP|CENB_HUMAN (P07199) Major centromere autoantigen B
(Centromere protein B) (CENP-B), partial (5%)
Length = 710
Score = 164 bits (414), Expect = 2e-41
Identities = 89/164 (54%), Positives = 119/164 (72%), Gaps = 11/164 (6%)
Frame = +1
Query: 14 LPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKSLADITDLLSTATEHR 73
LP LMIEIL + +NPLQLR VCK WKSLVVD QFV+KHLH SL+DITDL S A E
Sbjct: 196 LPEGLMIEILVWIRVSNPLQLRCVCKRWKSLVVDPQFVKKHLHTSLSDITDLASNAMEDM 375
Query: 74 KSF---------VSHQIPNQLQEEEEEDDDDDDDDEEVDEEEAAKRAR--MNRLAELDNL 122
+F ++ + +EEEEE++++++++EE EEE + AR +N LA+LDN+
Sbjct: 376 NAFQL*LNYAPALAEPEEEEEEEEEEEEENEEEEEEEEGEEEEEEEARSMVNELAQLDNM 555
Query: 123 LVGVRSIKRELEIIKVDTLAMKDRMKCLESFLKIYLKSATSSSS 166
LV +RS+K LE IKVD A+K+R+KCL+SFL+IYLK+AT+SSS
Sbjct: 556 LVVIRSLKGTLETIKVDVQAIKERVKCLQSFLQIYLKTATASSS 687
>AI443156
Length = 414
Score = 52.0 bits (123), Expect = 1e-07
Identities = 24/58 (41%), Positives = 37/58 (63%)
Frame = +3
Query: 1 MMADATAESPTTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKS 58
M A+ S LPVE++ EIL+R+ + ++LRS CKWW+S++ F+ HL+KS
Sbjct: 51 MHANPITVSNMANLPVEVVTEILSRLPVKSVIRLRSTCKWWRSIIDSRHFILFHLNKS 224
>TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, partial (8%)
Length = 1061
Score = 51.6 bits (122), Expect = 2e-07
Identities = 22/45 (48%), Positives = 33/45 (72%)
Frame = +3
Query: 14 LPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKS 58
LPVE++ EIL+R+ + ++LRS CKWW+S++ FV HL+KS
Sbjct: 93 LPVEVVTEILSRLPVKSVIRLRSTCKWWRSIIDSRHFVLFHLNKS 227
>TC219823 weakly similar to UP|Q7RJ53 (Q7RJ53) Arabidopsis thaliana
At4g22410/F7K2_7-related, partial (3%)
Length = 613
Score = 45.4 bits (106), Expect(2) = 2e-06
Identities = 22/34 (64%), Positives = 28/34 (81%)
Frame = +3
Query: 113 MNRLAELDNLLVGVRSIKRELEIIKVDTLAMKDR 146
MN LA+LDN+LV VRS+K LE+IKVD A+K+R
Sbjct: 84 MNELAQLDNILVVVRSLKGSLEMIKVDVQAIKER 185
Score = 21.9 bits (45), Expect(2) = 2e-06
Identities = 10/17 (58%), Positives = 11/17 (63%)
Frame = +1
Query: 60 ADITDLLSTATEHRKSF 76
ADITDL S A E +F
Sbjct: 1 ADITDLASNAMEDMNAF 51
>AW395660
Length = 381
Score = 43.5 bits (101), Expect = 5e-05
Identities = 20/43 (46%), Positives = 28/43 (64%)
Frame = +3
Query: 10 PTTILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQ 52
P LP EL++EIL+R+ + LQ R VCK W SL+ D F++
Sbjct: 252 PLPFLPDELVVEILSRLPVKSLLQFRCVCKSWMSLIYDPYFMK 380
>TC232958
Length = 758
Score = 41.2 bits (95), Expect = 2e-04
Identities = 25/64 (39%), Positives = 39/64 (60%), Gaps = 3/64 (4%)
Frame = +1
Query: 13 ILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKH--LHKSLAD-ITDLLSTA 69
+LP +L+ EIL R+ + ++ +SVCK W L+ D +F + H L +LAD I + S+A
Sbjct: 58 VLPQDLITEILLRLPVKSLVRFKSVCKSWLFLISDPRFAKSHFDLFAALADRILFIASSA 237
Query: 70 TEHR 73
E R
Sbjct: 238 PELR 249
>TC215451 weakly similar to UP|Q94K07 (Q94K07) Nucleosome assembly protein,
partial (20%)
Length = 777
Score = 38.9 bits (89), Expect = 0.001
Identities = 15/33 (45%), Positives = 26/33 (78%)
Frame = +2
Query: 87 EEEEEDDDDDDDDEEVDEEEAAKRARMNRLAEL 119
++E+E+DDDDDDD++ DEEE+ + ++ +L L
Sbjct: 125 DDEDEEDDDDDDDDDDDEEESKTKKKV*QLMSL 223
Score = 35.0 bits (79), Expect = 0.017
Identities = 11/24 (45%), Positives = 22/24 (90%)
Frame = +2
Query: 86 QEEEEEDDDDDDDDEEVDEEEAAK 109
++++E+++DDDDDD++ D+EE +K
Sbjct: 119 EDDDEDEEDDDDDDDDDDDEEESK 190
Score = 34.7 bits (78), Expect = 0.022
Identities = 10/28 (35%), Positives = 23/28 (81%)
Frame = +2
Query: 85 LQEEEEEDDDDDDDDEEVDEEEAAKRAR 112
++E+++ED++DDDDD++ D++E + +
Sbjct: 113 IEEDDDEDEEDDDDDDDDDDDEEESKTK 196
Score = 28.5 bits (62), Expect = 1.6
Identities = 9/20 (45%), Positives = 18/20 (90%)
Frame = +2
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
E++E+ ++DDD+DEE D+++
Sbjct: 98 EDDEDIEEDDDEDEEDDDDD 157
Score = 26.9 bits (58), Expect = 4.6
Identities = 9/22 (40%), Positives = 18/22 (80%)
Frame = +2
Query: 85 LQEEEEEDDDDDDDDEEVDEEE 106
+Q +E D +DD+DDE+++E++
Sbjct: 62 IQGDEFGDLEDDEDDEDIEEDD 127
>TC215450 similar to UP|P93488 (P93488) NAP1Ps, partial (30%)
Length = 672
Score = 38.9 bits (89), Expect = 0.001
Identities = 17/41 (41%), Positives = 29/41 (70%)
Frame = +3
Query: 87 EEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELDNLLVGVR 127
++EEE++D+DDDD+E DEEE+ + + + A+L + G R
Sbjct: 246 DDEEEEEDEDDDDDEDDEEESKTKKKKSGRAQLGDGQQGER 368
Score = 37.0 bits (84), Expect = 0.004
Identities = 15/27 (55%), Positives = 20/27 (73%)
Frame = +3
Query: 86 QEEEEEDDDDDDDDEEVDEEEAAKRAR 112
+EEE+EDDDDD+DDEE + + K R
Sbjct: 255 EEEEDEDDDDDEDDEEESKTKKKKSGR 335
Score = 30.8 bits (68), Expect = 0.32
Identities = 10/22 (45%), Positives = 20/22 (90%)
Frame = +3
Query: 85 LQEEEEEDDDDDDDDEEVDEEE 106
L+++E+++D ++DDDEE +E+E
Sbjct: 207 LEDDEDDEDIEEDDDEEEEEDE 272
Score = 28.5 bits (62), Expect = 1.6
Identities = 9/20 (45%), Positives = 18/20 (90%)
Frame = +3
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
E++E+ ++DDD++EE DE++
Sbjct: 219 EDDEDIEEDDDEEEEEDEDD 278
Score = 28.5 bits (62), Expect = 1.6
Identities = 9/20 (45%), Positives = 18/20 (90%)
Frame = +3
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
+E++ED ++DDD+EE ++E+
Sbjct: 216 DEDDEDIEEDDDEEEEEDED 275
Score = 26.9 bits (58), Expect = 4.6
Identities = 9/22 (40%), Positives = 18/22 (80%)
Frame = +3
Query: 85 LQEEEEEDDDDDDDDEEVDEEE 106
+Q +E D +DD+DDE+++E++
Sbjct: 183 IQGDEFGDLEDDEDDEDIEEDD 248
>TC233794 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {Arabidopsis
thaliana;} , partial (9%)
Length = 449
Score = 38.1 bits (87), Expect = 0.002
Identities = 20/43 (46%), Positives = 27/43 (62%)
Frame = +1
Query: 12 TILPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKH 54
T LP EL+ EIL R+ + L+ + VCK + SL+ D QFV H
Sbjct: 103 TTLPQELIREILLRLPVKSLLRFKCVCKSFLSLISDPQFVISH 231
>TC204848 similar to UP|Q6ICZ8 (Q6ICZ8) At5g13850, partial (89%)
Length = 916
Score = 37.7 bits (86), Expect = 0.003
Identities = 18/66 (27%), Positives = 35/66 (52%)
Frame = +3
Query: 64 DLLSTATEHRKSFVSHQIPNQLQEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELDNLL 123
+LL+ E +K + E++E+DDD+DDD+ E D +K+ R + + L
Sbjct: 99 ELLAADLEQQKIHDDEPVVEDDDEDDEDDDDEDDDNVEGDASGRSKQTRSEKKSRKAMLK 278
Query: 124 VGVRSI 129
+G++ +
Sbjct: 279 LGMKPV 296
>TC213957 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {Arabidopsis
thaliana;} , partial (9%)
Length = 696
Score = 37.7 bits (86), Expect = 0.003
Identities = 20/45 (44%), Positives = 26/45 (57%)
Frame = +3
Query: 14 LPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKS 58
LP+EL+ EIL R + L+ + VCK W SL+ D QF L S
Sbjct: 33 LPLELIREILLRSPVRSVLRFKCVCKSWLSLISDPQFTHFDLAAS 167
>TC215452 similar to UP|P93488 (P93488) NAP1Ps, partial (29%)
Length = 609
Score = 37.7 bits (86), Expect = 0.003
Identities = 14/24 (58%), Positives = 20/24 (83%)
Frame = +3
Query: 87 EEEEEDDDDDDDDEEVDEEEAAKR 110
+E+EEDDDDDDDD++ +EE K+
Sbjct: 177 DEDEEDDDDDDDDDDDEEESKTKK 248
Score = 37.0 bits (84), Expect = 0.004
Identities = 13/26 (50%), Positives = 22/26 (84%)
Frame = +3
Query: 87 EEEEEDDDDDDDDEEVDEEEAAKRAR 112
++E+E+DDDDDDD++ DEEE+ + +
Sbjct: 174 DDEDEEDDDDDDDDDDDEEESKTKKK 251
Score = 37.0 bits (84), Expect = 0.004
Identities = 13/25 (52%), Positives = 21/25 (84%)
Frame = +3
Query: 87 EEEEEDDDDDDDDEEVDEEEAAKRA 111
E+EE+DDDDDDDD++ +E + K++
Sbjct: 180 EDEEDDDDDDDDDDDEEESKTKKKS 254
Score = 35.0 bits (79), Expect = 0.017
Identities = 11/24 (45%), Positives = 22/24 (90%)
Frame = +3
Query: 86 QEEEEEDDDDDDDDEEVDEEEAAK 109
++++E+++DDDDDD++ D+EE +K
Sbjct: 168 EDDDEDEEDDDDDDDDDDDEEESK 239
Score = 34.7 bits (78), Expect = 0.022
Identities = 10/28 (35%), Positives = 23/28 (81%)
Frame = +3
Query: 85 LQEEEEEDDDDDDDDEEVDEEEAAKRAR 112
++E+++ED++DDDDD++ D++E + +
Sbjct: 162 IEEDDDEDEEDDDDDDDDDDDEEESKTK 245
Score = 33.9 bits (76), Expect = 0.038
Identities = 12/24 (50%), Positives = 19/24 (79%)
Frame = +3
Query: 86 QEEEEEDDDDDDDDEEVDEEEAAK 109
++EE++DDDDDDDD+E + + K
Sbjct: 180 EDEEDDDDDDDDDDDEEESKTKKK 251
Score = 33.1 bits (74), Expect = 0.064
Identities = 14/42 (33%), Positives = 27/42 (63%)
Frame = +3
Query: 86 QEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELDNLLVGVR 127
+E++++DDDDDDD+EE ++ + + + A+L + G R
Sbjct: 186 EEDDDDDDDDDDDEEESKTKKKSSAPKKSGRAQLGDGQQGER 311
Score = 28.5 bits (62), Expect = 1.6
Identities = 9/20 (45%), Positives = 18/20 (90%)
Frame = +3
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
E++E+ ++DDD+DEE D+++
Sbjct: 147 EDDEDIEEDDDEDEEDDDDD 206
Score = 28.1 bits (61), Expect = 2.1
Identities = 9/19 (47%), Positives = 17/19 (89%)
Frame = +3
Query: 88 EEEEDDDDDDDDEEVDEEE 106
E++EDD+D ++D++ DEE+
Sbjct: 138 EDDEDDEDIEEDDDEDEED 194
Score = 27.7 bits (60), Expect = 2.7
Identities = 8/20 (40%), Positives = 18/20 (90%)
Frame = +3
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
+E++ED ++DDD++E D+++
Sbjct: 144 DEDDEDIEEDDDEDEEDDDD 203
Score = 27.3 bits (59), Expect = 3.5
Identities = 7/22 (31%), Positives = 20/22 (90%)
Frame = +3
Query: 85 LQEEEEEDDDDDDDDEEVDEEE 106
L+++E+++D ++DDDE+ ++++
Sbjct: 135 LEDDEDDEDIEEDDDEDEEDDD 200
Score = 27.3 bits (59), Expect = 3.5
Identities = 16/60 (26%), Positives = 30/60 (49%), Gaps = 4/60 (6%)
Frame = +3
Query: 51 VQKHLHKSLADITDLLSTATEHRKSFVSHQIP----NQLQEEEEEDDDDDDDDEEVDEEE 106
V + L + D+ ST R + H + +Q +E D +DD+DDE+++E++
Sbjct: 6 VAEELQNQMEQDYDIGSTI---RDKIIPHAVSWFTGEAIQGDEFGDLEDDEDDEDIEEDD 176
Score = 26.9 bits (58), Expect = 4.6
Identities = 7/21 (33%), Positives = 19/21 (90%)
Frame = +3
Query: 86 QEEEEEDDDDDDDDEEVDEEE 106
+++E+ ++DDD+D+E+ D+++
Sbjct: 147 EDDEDIEEDDDEDEEDDDDDD 209
>TC215449 similar to UP|P93488 (P93488) NAP1Ps, partial (97%)
Length = 1495
Score = 37.0 bits (84), Expect = 0.004
Identities = 14/24 (58%), Positives = 20/24 (83%)
Frame = +2
Query: 87 EEEEEDDDDDDDDEEVDEEEAAKR 110
+EEEE+D+DDDDDE+ +EE K+
Sbjct: 1082 DEEEEEDEDDDDDEDDEEESKTKK 1153
Score = 36.2 bits (82), Expect = 0.008
Identities = 13/26 (50%), Positives = 22/26 (84%)
Frame = +2
Query: 87 EEEEEDDDDDDDDEEVDEEEAAKRAR 112
++EEE++D+DDDD+E DEEE+ + +
Sbjct: 1079 DDEEEEEDEDDDDDEDDEEESKTKKK 1156
Score = 36.2 bits (82), Expect = 0.008
Identities = 13/25 (52%), Positives = 21/25 (84%)
Frame = +2
Query: 87 EEEEEDDDDDDDDEEVDEEEAAKRA 111
EEEEED+DDDDD+++ +E + K++
Sbjct: 1085 EEEEEDEDDDDDEDDEEESKTKKKS 1159
Score = 34.7 bits (78), Expect = 0.022
Identities = 13/16 (81%), Positives = 16/16 (99%)
Frame = +2
Query: 86 QEEEEEDDDDDDDDEE 101
+EEE+EDDDDD+DDEE
Sbjct: 1088 EEEEDEDDDDDEDDEE 1135
Score = 33.9 bits (76), Expect = 0.038
Identities = 15/42 (35%), Positives = 27/42 (63%)
Frame = +2
Query: 86 QEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELDNLLVGVR 127
+EE+E+DDDD+DD+EE ++ + + + A+L + G R
Sbjct: 1091 EEEDEDDDDDEDDEEESKTKKKSSAPKKSGRAQLGDGQQGER 1216
Score = 33.5 bits (75), Expect = 0.049
Identities = 10/24 (41%), Positives = 22/24 (91%)
Frame = +2
Query: 86 QEEEEEDDDDDDDDEEVDEEEAAK 109
++++EE+++D+DDD++ D+EE +K
Sbjct: 1073 EDDDEEEEEDEDDDDDEDDEEESK 1144
Score = 33.1 bits (74), Expect = 0.064
Identities = 12/24 (50%), Positives = 19/24 (79%)
Frame = +2
Query: 86 QEEEEEDDDDDDDDEEVDEEEAAK 109
+EEEE++DDDDD+D+E + + K
Sbjct: 1085 EEEEEDEDDDDDEDDEEESKTKKK 1156
Score = 30.8 bits (68), Expect = 0.32
Identities = 10/22 (45%), Positives = 20/22 (90%)
Frame = +2
Query: 85 LQEEEEEDDDDDDDDEEVDEEE 106
L+++E+++D ++DDDEE +E+E
Sbjct: 1040 LEDDEDDEDIEEDDDEEEEEDE 1105
Score = 29.3 bits (64), Expect = 0.93
Identities = 17/63 (26%), Positives = 32/63 (49%), Gaps = 4/63 (6%)
Frame = +2
Query: 48 DQFVQKHLHKSLADITDLLSTATEHRKSFVSHQIP----NQLQEEEEEDDDDDDDDEEVD 103
D+ V + L + D+ ST R + H + +Q +E D +DD+DDE+++
Sbjct: 902 DEDVAEELQNQMEQDYDIGSTI---RDKIIPHAVSWFTGEAIQGDEFGDLEDDEDDEDIE 1072
Query: 104 EEE 106
E++
Sbjct: 1073EDD 1081
Score = 28.5 bits (62), Expect = 1.6
Identities = 9/20 (45%), Positives = 18/20 (90%)
Frame = +2
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
E++E+ ++DDD++EE DE++
Sbjct: 1052 EDDEDIEEDDDEEEEEDEDD 1111
Score = 28.5 bits (62), Expect = 1.6
Identities = 9/20 (45%), Positives = 18/20 (90%)
Frame = +2
Query: 87 EEEEEDDDDDDDDEEVDEEE 106
+E++ED ++DDD+EE ++E+
Sbjct: 1049 DEDDEDIEEDDDEEEEEDED 1108
Score = 27.3 bits (59), Expect = 3.5
Identities = 21/83 (25%), Positives = 40/83 (47%), Gaps = 3/83 (3%)
Frame = +2
Query: 26 VDSNNPLQLRSV---CKWWKSLVVDDQFVQKHLHKSLADITDLLSTATEHRKSFVSHQIP 82
+D + P+ +++ +W + + ++K K + + T TE+ +SF + P
Sbjct: 698 IDEDEPILEKAIGTEIQWHPGKCLTQKILKKKPKKGSKNAKPI--TKTENCESFFNFFNP 871
Query: 83 NQLQEEEEEDDDDDDDDEEVDEE 105
Q+ E DD+D DE+V EE
Sbjct: 872 PQVPE------DDEDIDEDVAEE 922
>TC221531 similar to UP|Q84N48 (Q84N48) CRS2-associated factor 2, partial
(5%)
Length = 922
Score = 36.6 bits (83), Expect = 0.006
Identities = 17/47 (36%), Positives = 31/47 (65%), Gaps = 3/47 (6%)
Frame = +1
Query: 63 TDLLSTATEHRK---SFVSHQIPNQLQEEEEEDDDDDDDDEEVDEEE 106
+D L T ++ K S+ S + + +EEEEED+D+D+D++ D+E+
Sbjct: 568 SDGLHTEEDYAKFEDSYSSDALQTEEEEEEEEDEDEDEDEDYYDDED 708
Score = 28.1 bits (61), Expect = 2.1
Identities = 12/27 (44%), Positives = 20/27 (73%), Gaps = 2/27 (7%)
Frame = +1
Query: 80 QIPNQLQEEEEEDDDDDDD--DEEVDE 104
Q + +EEE+ED+D+D+D D+E D+
Sbjct: 634 QTEEEEEEEEDEDEDEDEDYYDDEDDD 714
>TC235094 similar to UP|PRT3_SCYCA (P30258) Protamine Z3 (Scylliorhinine Z3),
partial (76%)
Length = 466
Score = 36.6 bits (83), Expect = 0.006
Identities = 27/87 (31%), Positives = 45/87 (51%), Gaps = 4/87 (4%)
Frame = -2
Query: 80 QIPNQLQEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELDNLLVGVRSIKRELEIIKVD 139
+I E+EEE++DDDDDD E+ R R R ++L+ L R + EL I+V
Sbjct: 342 EIDEPEDEDEEEEEDDDDDDVVSQEQSPLSRLREQR-SKLETL---SRRLASELVPIRVH 175
Query: 140 TLAMKDRMK----CLESFLKIYLKSAT 162
+ ++ K +E+ LK+ ++ T
Sbjct: 174 DVLIRGNTKTKAWLIEAELKLLEEATT 94
>TC203654 similar to PIR|T46225|T46225 alpha NAC-like protein - Arabidopsis
thaliana {Arabidopsis thaliana;} , partial (76%)
Length = 1139
Score = 36.6 bits (83), Expect = 0.006
Identities = 17/47 (36%), Positives = 30/47 (63%), Gaps = 3/47 (6%)
Frame = +3
Query: 86 QEEEEEDDDDDDDDEEVDEE---EAAKRARMNRLAELDNLLVGVRSI 129
+E E+ED+DDDDDD+E D + E K++R + + L +G++ +
Sbjct: 234 EETEDEDEDDDDDDKEDDAQGGAEGGKQSRSEKKSRKAMLKLGLKPV 374
>BG404995 similar to GP|22831068|dbj OJ1131_E05.32 {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 394
Score = 36.2 bits (82), Expect = 0.008
Identities = 19/46 (41%), Positives = 27/46 (58%)
Frame = +1
Query: 14 LPVELMIEILARVDSNNPLQLRSVCKWWKSLVVDDQFVQKHLHKSL 59
LP E++ EIL+R+ + L+ RS K WKSL+ HL +SL
Sbjct: 40 LPREVLTEILSRLPVRSLLRFRSTSKSWKSLIDSQHLNWLHLTRSL 177
>BI320752
Length = 151
Score = 35.4 bits (80), Expect = 0.013
Identities = 15/34 (44%), Positives = 21/34 (61%)
Frame = +1
Query: 88 EEEEDDDDDDDDEEVDEEEAAKRARMNRLAELDN 121
EE +DDD+DDDD+E D+ E +EL+N
Sbjct: 1 EESDDDDEDDDDDEADDTEVGGLVGDGTKSELNN 102
Score = 31.2 bits (69), Expect = 0.24
Identities = 12/19 (63%), Positives = 15/19 (78%)
Frame = +1
Query: 87 EEEEEDDDDDDDDEEVDEE 105
EE ++DD+DDDDDE D E
Sbjct: 1 EESDDDDEDDDDDEADDTE 57
Score = 26.6 bits (57), Expect = 6.0
Identities = 9/16 (56%), Positives = 14/16 (87%)
Frame = +1
Query: 87 EEEEEDDDDDDDDEEV 102
+++E+DDDD+ DD EV
Sbjct: 13 DDDEDDDDDEADDTEV 60
>BU551036
Length = 340
Score = 35.4 bits (80), Expect = 0.013
Identities = 15/37 (40%), Positives = 24/37 (64%)
Frame = +2
Query: 85 LQEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELDN 121
+ E+E+EDDD+DD++EE ++E+A E DN
Sbjct: 41 VDEDEDEDDDEDDENEEYEDEDAFDAHAHASAGEHDN 151
Score = 27.7 bits (60), Expect = 2.7
Identities = 10/24 (41%), Positives = 17/24 (70%)
Frame = +2
Query: 83 NQLQEEEEEDDDDDDDDEEVDEEE 106
+ L + + D+D+D+DD+E DE E
Sbjct: 17 DDLHDGTDVDEDEDEDDDEDDENE 88
>BU761594 similar to GP|22654971|gb At1g80930/F23A5_23 {Arabidopsis
thaliana}, partial (13%)
Length = 397
Score = 35.4 bits (80), Expect = 0.013
Identities = 20/57 (35%), Positives = 31/57 (54%)
Frame = +1
Query: 71 EHRKSFVSHQIPNQLQEEEEEDDDDDDDDEEVDEEEAAKRARMNRLAELDNLLVGVR 127
E +KS + + + + + E DDDDD+D+E DEEE +M E + LV +R
Sbjct: 163 ELKKSMLGEESEDDEEGLDSESDDDDDEDDESDEEE---EEQMQIKDETETNLVNLR 324
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.129 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,140,947
Number of Sequences: 63676
Number of extensions: 78258
Number of successful extensions: 3949
Number of sequences better than 10.0: 340
Number of HSP's better than 10.0 without gapping: 1219
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2400
length of query: 168
length of database: 12,639,632
effective HSP length: 90
effective length of query: 78
effective length of database: 6,908,792
effective search space: 538885776
effective search space used: 538885776
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC149207.4


