

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149197.3 - phase: 0
(257 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
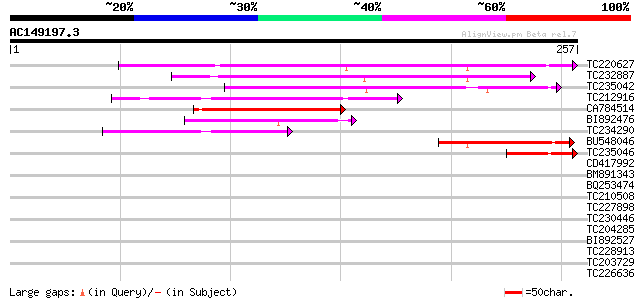
Score E
Sequences producing significant alignments: (bits) Value
TC220627 similar to UP|Q9LI79 (Q9LI79) Gb|AAB84348.1, partial (48%) 151 3e-37
TC232887 127 5e-30
TC235042 117 4e-27
TC212916 similar to UP|Q9FKQ4 (Q9FKQ4) Gb|AAD22354.1 (At5g65340)... 89 3e-18
CA784514 67 1e-11
BI892476 54 9e-08
TC234290 weakly similar to UP|Q9FL47 (Q9FL47) Gb|AAD20416.1, par... 52 2e-07
BU548046 43 2e-04
TC235046 43 2e-04
CD417992 38 0.004
BM891343 37 0.007
BQ253474 36 0.020
TC210508 36 0.020
TC227898 similar to GB|AAM19935.1|20453393|AY097419 AT5g58640/mz... 35 0.026
TC230446 weakly similar to UP|Q6V9S3 (Q6V9S3) RPE1 protein, part... 35 0.044
TC204285 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 34 0.057
BI892527 34 0.075
TC228913 weakly similar to UP|Q8IP68 (Q8IP68) CG31813-PA, partia... 34 0.075
TC203729 homologue to UP|Q9XQB4 (Q9XQB4) Photosystem I reaction ... 34 0.075
TC226636 UP|4CL2_SOYBN (P31687) 4-coumarate--CoA ligase 2 (4CL ... 34 0.075
>TC220627 similar to UP|Q9LI79 (Q9LI79) Gb|AAB84348.1, partial (48%)
Length = 1053
Score = 151 bits (381), Expect = 3e-37
Identities = 82/216 (37%), Positives = 130/216 (59%), Gaps = 8/216 (3%)
Frame = +2
Query: 50 SKQNSTNKLFGKFRSMFRSFPIIVPSCKMPTMNGNHRTSETIIHGGIRITGTLFGYRKAR 109
S+ N+ K +S F+S I ++P + N+ ++ + +R+ GTLFG+R+
Sbjct: 332 SQYNAKKKTHFSKKSKFKSVLTIFTVTRVPRLRLNNISNSFNLD--MRVMGTLFGHRRDH 505
Query: 110 VNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECE-KHSSNDKTKIVDEPIWT 168
V AFQED K P L++LA T L+++M GL RIALECE K ++ K ++++EP+W
Sbjct: 506 VYFAFQEDPKLGPTFLVKLATRTSTLVREMASGLVRIALECEKKKNATAKVRLLEEPLWR 685
Query: 169 LFCNGKKMGYGVKRDPTDDDLYVIQMLHSVSVAVGELP-------SDMSDPQDGELSYMR 221
+CNG+K GY +R+ + +++ + +S+ G LP + + GE+ YMR
Sbjct: 686 TYCNGRKCGYANRRECGPHEWKILKAVEPISMGAGVLPVVCGNEAAGSEEEASGEVMYMR 865
Query: 222 AHFERVIGSKDSETYYMMMPDGNSNGPELSVFFVRV 257
A +ERV+GS+DSE +YMM PD S GPELS++ +RV
Sbjct: 866 AKYERVVGSRDSEAFYMMNPDA-SGGPELSIYLIRV 970
>TC232887
Length = 555
Score = 127 bits (319), Expect = 5e-30
Identities = 73/187 (39%), Positives = 104/187 (55%), Gaps = 22/187 (11%)
Frame = +2
Query: 74 PSCKMPTMNGNHRTSETIIHGGIRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTG 133
P+CK T+ + ++ G ++TGTLFG+R+ ++ A Q + P LLLELA+ T
Sbjct: 2 PTCKWLTIPSQLSVTPSL---GRKVTGTLFGHRRGHISFAVQLHPRADPVLLLELAMSTS 172
Query: 134 KLLQDMGMGLNRIALECEKHSSNDKT-------------KIVDEPIWTLFCNGKKMGYGV 180
L+++M GL RIALE +K S++ T K+ EP WT++CNG+ GY V
Sbjct: 173 SLVKEMSSGLVRIALESQKLSASTITRTMRSNSGRQQQCKLFQEPSWTMYCNGRNCGYAV 352
Query: 181 KRDPTDDDLYVIQMLHSVSVAVGELP---------SDMSDPQDGELSYMRAHFERVIGSK 231
R D D +V+ + SVSV G +P + +GEL YMRA FERV+GS+
Sbjct: 353 SRTCGDLDWHVLSTIQSVSVGAGVIPLLEDGKAASAAAGGGSEGELMYMRARFERVVGSR 532
Query: 232 DSETYYM 238
DSE YM
Sbjct: 533 DSEAXYM 553
>TC235042
Length = 1174
Score = 117 bits (294), Expect = 4e-27
Identities = 64/160 (40%), Positives = 93/160 (58%), Gaps = 7/160 (4%)
Frame = +1
Query: 98 ITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECEKHSSND 157
+ GTLFGYR+ V+ AFQ+D P L+ELA P L+++M GL RIALEC+K ++
Sbjct: 448 LLGTLFGYRRGHVHFAFQKDPTSQPAFLIELATPISGLVREMASGLVRIALECDKDRDSE 627
Query: 158 KTK---IVDEPIWTLFCNGKKMGYGVKRDPTDDDLYVIQMLHSVSVAVGELPSDMSDPQD 214
K K ++ E +W +CNGKK G+ +R+ D +++ + +S+ G LP+ D
Sbjct: 628 KKKTLRLLQESVWRTYCNGKKCGFATRRECGAKDWDILKAVEPISMGAGVLPN-----SD 792
Query: 215 G----ELSYMRAHFERVIGSKDSETYYMMMPDGNSNGPEL 250
G + FER+ GS+DSE +YMM PD N PEL
Sbjct: 793 GARW*KSCT*XQXFERIXGSRDSEAFYMMNPDSN-GAPEL 909
>TC212916 similar to UP|Q9FKQ4 (Q9FKQ4) Gb|AAD22354.1 (At5g65340), partial
(12%)
Length = 446
Score = 88.6 bits (218), Expect = 3e-18
Identities = 50/132 (37%), Positives = 77/132 (57%)
Frame = +1
Query: 47 NSKSKQNSTNKLFGKFRSMFRSFPIIVPSCKMPTMNGNHRTSETIIHGGIRITGTLFGYR 106
N +SK +S L M + FP++ CKM + G R + TGT+FGYR
Sbjct: 76 NKRSKLSSGGGLL----KMLKLFPMLSSGCKMVALLGRPRK----MLKDSATTGTIFGYR 231
Query: 107 KARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGLNRIALECEKHSSNDKTKIVDEPI 166
K RV+LA QED++ P L+EL + L ++M + RIALE E + ++K K+++E +
Sbjct: 232 KGRVSLAIQEDTRQMPIFLIELPMMASALNKEMASDIVRIALESE--TKSNKKKLLEEFV 405
Query: 167 WTLFCNGKKMGY 178
W ++CNG+K+GY
Sbjct: 406 WAVYCNGRKVGY 441
>CA784514
Length = 435
Score = 66.6 bits (161), Expect = 1e-11
Identities = 35/69 (50%), Positives = 43/69 (61%)
Frame = +2
Query: 84 NHRTSETIIHGGIRITGTLFGYRKARVNLAFQEDSKCHPFLLLELAIPTGKLLQDMGMGL 143
NHR S G R+ GTLFGYR+ V+ AFQ D P L+ELA P L+++M GL
Sbjct: 227 NHR-SNLPFGLGSRVVGTLFGYRRGHVHFAFQRDPTSQPAFLIELATPISGLVREMASGL 403
Query: 144 NRIALECEK 152
RIALEC+K
Sbjct: 404 VRIALECDK 430
>BI892476
Length = 406
Score = 53.5 bits (127), Expect = 9e-08
Identities = 32/79 (40%), Positives = 46/79 (57%), Gaps = 1/79 (1%)
Frame = +3
Query: 80 TMNGNHRTSETIIHGGIRITGTLFGYRKARVNLAFQEDSKC-HPFLLLELAIPTGKLLQD 138
T + S +++ ITGT+FGYR+ +V+ Q ++ +P LLLELA+PT L ++
Sbjct: 171 TSKYTYSPSSSLMSSTTNITGTIFGYRRGKVSFCIQANANSTNPILLLELAVPTTILAKE 350
Query: 139 MGMGLNRIALECEKHSSND 157
M G RIALE S ND
Sbjct: 351 MRGGTLRIALE----SGND 395
>TC234290 weakly similar to UP|Q9FL47 (Q9FL47) Gb|AAD20416.1, partial (11%)
Length = 454
Score = 52.4 bits (124), Expect = 2e-07
Identities = 30/86 (34%), Positives = 46/86 (52%)
Frame = +1
Query: 43 LQPANSKSKQNSTNKLFGKFRSMFRSFPIIVPSCKMPTMNGNHRTSETIIHGGIRITGTL 102
L+ ++K+++ G M + FP++ CKM + G R + TGT+
Sbjct: 193 LRHTTGENKRSNKLSSGGGLLKMLKLFPMLSSGCKMVALLGRPRK----MLKDSATTGTI 360
Query: 103 FGYRKARVNLAFQEDSKCHPFLLLEL 128
FGYRK RV+LA QED++ P L+EL
Sbjct: 361 FGYRKGRVSLAIQEDTRQMPVFLIEL 438
>BU548046
Length = 195
Score = 42.7 bits (99), Expect = 2e-04
Identities = 24/63 (38%), Positives = 40/63 (63%), Gaps = 1/63 (1%)
Frame = -2
Query: 195 LHSVSVAVGELP-SDMSDPQDGELSYMRAHFERVIGSKDSETYYMMMPDGNSNGPELSVF 253
+ SV V G + ++ + +D +L Y+RA F+RV GS + E++ + P+G+ + ELSVF
Sbjct: 188 MRSVVVGTGVMKCKELVNKEDDQLMYLRASFQRVRGSDNCESFXXIDPEGDID-QELSVF 12
Query: 254 FVR 256
F R
Sbjct: 11 FFR 3
>TC235046
Length = 459
Score = 42.7 bits (99), Expect = 2e-04
Identities = 18/32 (56%), Positives = 25/32 (77%)
Frame = +2
Query: 226 RVIGSKDSETYYMMMPDGNSNGPELSVFFVRV 257
R++GS+DSE +YMM PD N PELS++ +RV
Sbjct: 14 RIVGSRDSEAFYMMNPDSN-GAPELSIYLLRV 106
>CD417992
Length = 138
Score = 38.1 bits (87), Expect = 0.004
Identities = 18/45 (40%), Positives = 29/45 (64%), Gaps = 1/45 (2%)
Frame = -2
Query: 181 KRDPTDDDLYVIQMLHSVSVAVGELPSDMSDPQ-DGELSYMRAHF 224
++ +DD+L+V+Q L VS+ G LP+ DGE++Y+RA F
Sbjct: 137 RKQMSDDELHVMQHLRGVSMGAGVLPTSSDHKDCDGEMTYIRASF 3
>BM891343
Length = 418
Score = 37.4 bits (85), Expect = 0.007
Identities = 22/51 (43%), Positives = 30/51 (58%)
Frame = +2
Query: 7 SQTTTGQPPPSPTTGQPPPSPASVNTCPPTVRMPINLQPANSKSKQNSTNK 57
S+TT P PS TT +P P+P S P+ P L+ ANS S+ NS++K
Sbjct: 8 SKTTPKSPSPSSTTPKPSPTPLSPTPPSPSSSTPW-LRFANSTSRGNSSSK 157
>BQ253474
Length = 276
Score = 35.8 bits (81), Expect = 0.020
Identities = 19/52 (36%), Positives = 26/52 (49%), Gaps = 4/52 (7%)
Frame = +3
Query: 10 TTGQPPPSPTT----GQPPPSPASVNTCPPTVRMPINLQPANSKSKQNSTNK 57
T PPPSP T G PPPS AS +T P + S ++ +++NK
Sbjct: 114 TVPSPPPSPATPSAYGMPPPSAASSSTPSAAAPAPATPRDPTSPTRNDTSNK 269
>TC210508
Length = 413
Score = 35.8 bits (81), Expect = 0.020
Identities = 21/75 (28%), Positives = 32/75 (42%)
Frame = +3
Query: 6 ESQTTTGQPPPSPTTGQPPPSPASVNTCPPTVRMPINLQPANSKSKQNSTNKLFGKFRSM 65
++ T++ PPPSPTT S S PP R P A S+ +++ +K + +
Sbjct: 3 QNSTSSSSPPPSPTTRT*ARS--SARPSPPASRKPSTTTSATSRGRKSQRSKRCARLTTR 176
Query: 66 FRSFPIIVPSCKMPT 80
S P PT
Sbjct: 177TSSSPSTTSDLSSPT 221
>TC227898 similar to GB|AAM19935.1|20453393|AY097419 AT5g58640/mzn1_90
{Arabidopsis thaliana;} , partial (77%)
Length = 1028
Score = 35.4 bits (80), Expect = 0.026
Identities = 18/37 (48%), Positives = 19/37 (50%), Gaps = 1/37 (2%)
Frame = +2
Query: 5 HESQTTTGQPP-PSPTTGQPPPSPASVNTCPPTVRMP 40
H S TT PP P P + PPP PAS T PP P
Sbjct: 185 HSSSTTATPPPSPPPPSTSPPPPPASPPTPPPPSPAP 295
Score = 29.6 bits (65), Expect = 1.4
Identities = 18/44 (40%), Positives = 21/44 (46%), Gaps = 2/44 (4%)
Frame = +2
Query: 7 SQTTTGQPPPSPTTGQPPPS--PASVNTCPPTVRMPINLQPANS 48
S +TT PPPSP PPPS P PPT P P ++
Sbjct: 188 SSSTTATPPPSP----PPPSTSPPPPPASPPTPPPPSPAPPPSA 307
Score = 29.3 bits (64), Expect = 1.8
Identities = 17/48 (35%), Positives = 25/48 (51%), Gaps = 3/48 (6%)
Frame = +2
Query: 9 TTTGQPPPSPTTGQPPPSPASVNTCPPTVRMP---INLQPANSKSKQN 53
+T+ PPP+ PPPSPA + R+P + Q NS S+Q+
Sbjct: 233 STSPPPPPASPPTPPPPSPAPPPSAAGDNRVPSRETHQQYCNSGSRQH 376
>TC230446 weakly similar to UP|Q6V9S3 (Q6V9S3) RPE1 protein, partial (12%)
Length = 803
Score = 34.7 bits (78), Expect = 0.044
Identities = 19/57 (33%), Positives = 32/57 (55%), Gaps = 5/57 (8%)
Frame = +2
Query: 5 HESQTTTGQPPPSPTTGQPPPSPASVNTCPPTVRMP--INLQPAN---SKSKQNSTN 56
HES + PP S ++ PPPS + ++ PP+ R P + PA+ +K+ +ST+
Sbjct: 2 HESASAPAPPPASSSSTTPPPSSTAGSSTPPSSRSPASTSTSPASRGTTKTASSSTS 172
>TC204285 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (32%)
Length = 1539
Score = 34.3 bits (77), Expect = 0.057
Identities = 15/36 (41%), Positives = 17/36 (46%)
Frame = +1
Query: 14 PPPSPTTGQPPPSPASVNTCPPTVRMPINLQPANSK 49
PPP P +PPP P CPP V P +P K
Sbjct: 253 PPPKPPPVKPPPKPPVKPPCPPKVEPPHPPKPPKVK 360
Score = 32.0 bits (71), Expect = 0.28
Identities = 15/38 (39%), Positives = 15/38 (39%)
Frame = +1
Query: 14 PPPSPTTGQPPPSPASVNTCPPTVRMPINLQPANSKSK 51
PPP P PPP P CPP V P P K
Sbjct: 178 PPPVPEKKPPPPVPEKPPPCPPKVVPPPKPPPVKPPPK 291
Score = 27.7 bits (60), Expect = 5.4
Identities = 12/28 (42%), Positives = 14/28 (49%), Gaps = 2/28 (7%)
Frame = +1
Query: 15 PPSPTTGQPP--PSPASVNTCPPTVRMP 40
PP P +PP P P CPP V+ P
Sbjct: 424 PPKPPKVEPPHPPKPPKKPPCPPKVKPP 507
Score = 26.9 bits (58), Expect = 9.2
Identities = 12/31 (38%), Positives = 17/31 (54%), Gaps = 2/31 (6%)
Frame = +1
Query: 13 QPPPSPTTG--QPPPSPASVNTCPPTVRMPI 41
+PP P T +PPP+P CPP + P+
Sbjct: 637 KPPKEPPTPPKEPPPTPPK-KPCPPIFKKPL 726
>BI892527
Length = 423
Score = 33.9 bits (76), Expect = 0.075
Identities = 19/46 (41%), Positives = 22/46 (47%)
Frame = +2
Query: 7 SQTTTGQPPPSPTTGQPPPSPASVNTCPPTVRMPINLQPANSKSKQ 52
S T+ PPPSP PPPSP S PP +QP +S Q
Sbjct: 131 STTSPSPPPPSP----PPPSPPSSYLNPPRRNNKARVQPLRLRSLQ 256
>TC228913 weakly similar to UP|Q8IP68 (Q8IP68) CG31813-PA, partial (19%)
Length = 692
Score = 33.9 bits (76), Expect = 0.075
Identities = 17/39 (43%), Positives = 22/39 (55%)
Frame = +2
Query: 9 TTTGQPPPSPTTGQPPPSPASVNTCPPTVRMPINLQPAN 47
TT+ PP SP + PP SP S +T PT P N P++
Sbjct: 53 TTSTLPPASPRSSSPPSSPPSPST-TPTSSAPANAPPSS 166
Score = 27.7 bits (60), Expect = 5.4
Identities = 14/28 (50%), Positives = 17/28 (60%)
Frame = +3
Query: 13 QPPPSPTTGQPPPSPASVNTCPPTVRMP 40
QPPP P +PPP P S+ P VR+P
Sbjct: 27 QPPPPP--DEPPPQP-SLRPHPARVRLP 101
>TC203729 homologue to UP|Q9XQB4 (Q9XQB4) Photosystem I reaction center
subunit III, partial (94%)
Length = 1140
Score = 33.9 bits (76), Expect = 0.075
Identities = 20/62 (32%), Positives = 25/62 (40%), Gaps = 7/62 (11%)
Frame = +2
Query: 14 PPPSPTTGQPPPSPASVNTCPPTVR-------MPINLQPANSKSKQNSTNKLFGKFRSMF 66
PPP P + PP+ + TCPPT R P + P S S + RS
Sbjct: 191 PPPQPPSSAAPPTTTTTTTCPPT*RRSPPRWPSPPSSSPPLSPPAPTSRGSPHARSRSSS 370
Query: 67 RS 68
RS
Sbjct: 371 RS 376
>TC226636 UP|4CL2_SOYBN (P31687) 4-coumarate--CoA ligase 2 (4CL 2)
(4-coumaroyl-CoA synthase 2) (Clone 4CL16) , complete
Length = 1973
Score = 33.9 bits (76), Expect = 0.075
Identities = 18/48 (37%), Positives = 29/48 (59%)
Frame = -2
Query: 11 TGQPPPSPTTGQPPPSPASVNTCPPTVRMPINLQPANSKSKQNSTNKL 58
+GQ PP P+ P P+PA PPT+ ++ PA S+S+ +++ KL
Sbjct: 985 SGQNPP-PSDSLPTPTPAGAPPSPPTLGAAVS-APAVSRSRTSASTKL 848
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,734,219
Number of Sequences: 63676
Number of extensions: 226801
Number of successful extensions: 7705
Number of sequences better than 10.0: 468
Number of HSP's better than 10.0 without gapping: 3824
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6437
length of query: 257
length of database: 12,639,632
effective HSP length: 95
effective length of query: 162
effective length of database: 6,590,412
effective search space: 1067646744
effective search space used: 1067646744
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC149197.3


