

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
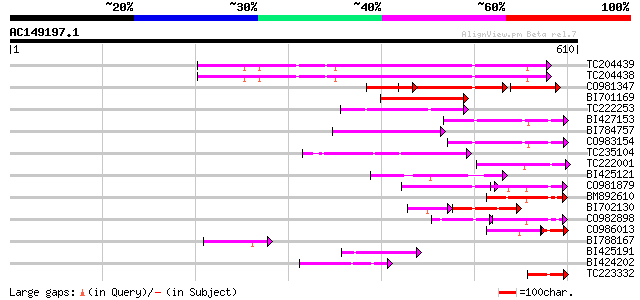
Query= AC149197.1 + phase: 0 /pseudo
(610 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 178 7e-45
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 176 2e-44
CO981347 111 2e-33
BI701169 99 5e-21
TC222253 similar to UP|Q9ZQE4 (Q9ZQE4) Copia-like retroelement p... 96 5e-20
BI427153 92 5e-19
BI784757 88 1e-17
CO983154 85 8e-17
TC235104 weakly similar to UP|Q850H8 (Q850H8) Gag-pol polyprotei... 84 1e-16
TC222001 similar to UP|C716_NEPRA (O04164) Cytochrome P450 71A6 ... 64 1e-10
BI425121 64 2e-10
CO981879 64 2e-10
BM892610 64 3e-10
BI702130 weakly similar to GP|4234854|gb| pol polyprotein {Zea m... 52 1e-09
CO982898 47 1e-08
CO986013 40 9e-07
BI788167 46 5e-05
BI425191 weakly similar to GP|14586969|gb| pol polyprotein {Citr... 46 5e-05
BI424202 45 9e-05
TC223332 44 2e-04
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 178 bits (451), Expect = 7e-45
Identities = 128/405 (31%), Positives = 199/405 (48%), Gaps = 24/405 (5%)
Frame = +1
Query: 203 LDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IG-IVHFKTND--------GV 253
LD+ + HM ++ EP V G+ + K+ +G +VH G+
Sbjct: 1690 LDSGCSRHMTGVKEFLLNIEPCSTSYVTFGDGSKGKIIGMGKLVHDGLPSLNKVLLVKGL 1869
Query: 254 VRTLTNESLVCKYS-----AEGGVLMVSKGYLVLLKASRI-DSLYVLQGIVVTGSAAVSS 307
L + S +C + L+ ++ VL+K SR D+ Y+ + S+ S
Sbjct: 1870 TANLISISQLCDEGFNVNFTKSECLVTNEKSEVLMKGSRSKDNCYLWTPQETSYSSTCLS 2049
Query: 308 SMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEF-----CKHYVFWKQKKVS 362
S K+D ++WH GH+ +GM + +G + +GI L+ C KQ K+S
Sbjct: 2050 S--KEDEVRIWHQRFGHLHLRGMKKIIDKGAV--RGIPNLKIEEGRICGECQIGKQVKMS 2217
Query: 363 FSTATHRTKG-ILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPT 421
H+T +L+ +H DL GP +V S GG+RY ++DDF R WV F+R K+ETF
Sbjct: 2218 HQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIREKSETFEV 2397
Query: 422 FKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERM 481
FK+ + ++ + +K++ +D+ EF +S F EFCT+ GI + PQQNG+ ER
Sbjct: 2398 FKELSLRLQREKDCVIKRIRSDHGREFENSRFTEFCTSEGITHEFSAAITPQQNGIVERK 2577
Query: 482 IRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYS 541
RTL E AR ML L +LW EA +TAC++ NR +IW G
Sbjct: 2578 NRTLQEAARVMLHAKEL--PYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPSVK 2751
Query: 542 NLRIFGCPAYALVN---DGKLAPRAVECIFLSYASESKGYRLWCS 583
+ IFG P Y L + K+ P++ IFL Y++ S+ YR++ S
Sbjct: 2752 HFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNS 2886
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 176 bits (447), Expect = 2e-44
Identities = 127/405 (31%), Positives = 198/405 (48%), Gaps = 24/405 (5%)
Frame = +1
Query: 203 LDT*YNFHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IG-IVHFKTND--------GV 253
LD+ + HM ++ EP V G+ + K+ +G +VH G+
Sbjct: 1693 LDSGCSRHMTGVKEFLVNIEPCSTSYVTFGDGSKGKITGMGKLVHDGLPSLNKVLLVKGL 1872
Query: 254 VRTLTNESLVCKYS-----AEGGVLMVSKGYLVLLKASRI-DSLYVLQGIVVTGSAAVSS 307
L + S +C + L+ ++ VL+K SR D+ Y+ + S+
Sbjct: 1873 TANLISISQLCDEGFNVNFTKSECLVTNEKSEVLMKGSRSKDNCYLWTPQETSYSSTCLF 2052
Query: 308 SMPKKDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEF-----CKHYVFWKQKKVS 362
S K+D K+WH GH+ +GM + +G + +GI L+ C KQ K+S
Sbjct: 2053 S--KEDEVKIWHQRFGHLHLRGMKKIIDKGAV--RGIPNLKIEEGRICGECQIGKQVKMS 2220
Query: 363 FSTATHRTKG-ILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPT 421
H+T +L+ +H DL GP +V S GG+RY ++DDF R WV F+R K++TF
Sbjct: 2221 HQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIREKSDTFEV 2400
Query: 422 FKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERM 481
FK+ + ++ + +K++ +D+ EF +S F EFCT+ GI + PQQNG+ ER
Sbjct: 2401 FKELSLRLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEFSAAITPQQNGIVERK 2580
Query: 482 IRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYS 541
RTL E AR ML L +LW EA +TAC++ NR +IW G
Sbjct: 2581 NRTLQEAARVMLHAKEL--PYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPTVK 2754
Query: 542 NLRIFGCPAYALVN---DGKLAPRAVECIFLSYASESKGYRLWCS 583
+ IFG P Y L + K+ P++ IFL Y++ S+ YR++ S
Sbjct: 2755 HFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNS 2889
>CO981347
Length = 624
Score = 111 bits (278), Expect(2) = 2e-33
Identities = 59/117 (50%), Positives = 72/117 (61%)
Frame = +2
Query: 419 FPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVA 478
F F++ L+ Q G +K L TDN LEF FNEFC GI RHK +P P QNG+A
Sbjct: 107 F*KFRE*HTLIGNQLGTKLKVLRTDNGLEFVLEQFNEFCRKIGIKRHKIVPHTP*QNGLA 286
Query: 479 ERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSG 535
ERM T+LER RCML +A L + W EAA+T +L+NR P S L FK P + WSG
Sbjct: 287 ERMNMTILERVRCMLLSARL--PKTFWGEAANTTSYLINRCPSSTLGFKTPMEAWSG 451
Score = 52.8 bits (125), Expect = 4e-07
Identities = 24/55 (43%), Positives = 37/55 (66%)
Frame = +3
Query: 384 PSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKWRILVETQTGKNVK 438
PS+V ++GG Y +TIIDDF R+VW+Y L+ K+E+F + +L+E +N K
Sbjct: 3 PSRVKTHGGSSYFLTIIDDFSRRVWLYVLKNKSESFKNSENDILLLEINLVQN*K 167
Score = 50.1 bits (118), Expect(2) = 2e-33
Identities = 22/54 (40%), Positives = 33/54 (60%)
Frame = +3
Query: 539 DYSNLRIFGCPAYALVNDGKLAPRAVECIFLSYASESKGYRLWCSDPKSQKLIL 592
+YS L++FG A+ V GKL RAV+C+F+ Y K Y+LW +P + I+
Sbjct: 462 NYSGLKVFGSLAFDHVKQGKLDARAVKCVFIGYPKGVKRYKLWKLEPGETRCII 623
>BI701169
Length = 407
Score = 99.0 bits (245), Expect = 5e-21
Identities = 47/94 (50%), Positives = 65/94 (69%)
Frame = +2
Query: 400 IDDFPRKVWVYFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTN 459
IDDF RK WVYF ++K E F FKK++ +VE ++G +K + +D EF S++F ++C +
Sbjct: 125 IDDFSRKTWVYFFKHKLEVFENFKKFKAIVEKESGFKIKAMRSDRGGEF*SNEFQKYCDD 304
Query: 460 HGIARHKTIPRNPQQNGVAERMIRTLLERARCML 493
HGI R + R+PQQNGVAER RT+L AR ML
Sbjct: 305 HGIRRPLMVLRSPQQNGVAERKNRTILNMARSML 406
>TC222253 similar to UP|Q9ZQE4 (Q9ZQE4) Copia-like retroelement pol
polyprotein, partial (4%)
Length = 919
Score = 95.9 bits (237), Expect = 5e-20
Identities = 54/138 (39%), Positives = 76/138 (54%), Gaps = 1/138 (0%)
Frame = +1
Query: 357 KQKKVSFSTA-THRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYK 415
K+ + SF T + R K +L +H DL ++ ++G Y +T IDDF +K+WVYFL+ K
Sbjct: 352 KKHRESFPTGKSWRMKKLLKIVHLDLC-TVEIPTHGDNNYFITFIDDFSKKMWVYFLKQK 528
Query: 416 NETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQN 475
+E FK ++ E Q G VK LI D E+ S + F HGI T PQ N
Sbjct: 529 SEACNAFKMFKAFAEKQNGCKVKALIIDKGQEYLS--YTIFFEKHGIQHQLTTKYTPQHN 702
Query: 476 GVAERMIRTLLERARCML 493
GV ER +T+++ RCML
Sbjct: 703 GVTERKNKTIMDMVRCML 756
>BI427153
Length = 422
Score = 92.4 bits (228), Expect = 5e-19
Identities = 55/141 (39%), Positives = 83/141 (58%), Gaps = 6/141 (4%)
Frame = +1
Query: 467 TIPRNPQQNGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDF 526
T P PQQNG+AER LLE AR ++ N+ + W +A TAC L+NR P S+L+
Sbjct: 10 TCPHTPQQNGIAERKNHHLLETARSLMLNSNVPT--HHWGDAVLTACFLINRMPSSSLEN 183
Query: 527 KVPEDI-WSGNLVDYSNLRIFGCPAYALVND-----GKLAPRAVECIFLSYASESKGYRL 580
++P I + +L+ Y + ++FGC + V+D KL+ R+V+C+FL Y+ KGY
Sbjct: 184 QIPHSIVFPNDLLFYVSPKVFGCTCF--VHDLSPGLDKLSARSVKCVFLGYSRLQKGYT- 354
Query: 581 WCSDPKSQKLILSRDVTFNED 601
C P ++ +S +VTF ED
Sbjct: 355 -CYFPNMRRYYMSANVTFFED 414
>BI784757
Length = 430
Score = 87.8 bits (216), Expect = 1e-17
Identities = 46/123 (37%), Positives = 70/123 (56%), Gaps = 1/123 (0%)
Frame = +1
Query: 348 EFCKHYVFWKQKKVSFS-TATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRK 406
E C + KQ + +F R K L+ I+SD+ GP + S GG RY ++ ID+ RK
Sbjct: 61 EVCDGCLQCKQSRSTFKQNVPIRAKEKLEVIYSDVCGPMQTESLGGNRYFISFIDELTRK 240
Query: 407 VWVYFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHK 466
VWVY +R K++ F F+K++ + + Q+G +K L T+ E+ S++F EFC GI
Sbjct: 241 VWVYLIRRKSDFFEVFEKFKNMAKKQSGSLIKILRTNGGGEYVSTEFQEFCDQQGIIHEV 420
Query: 467 TIP 469
T P
Sbjct: 421 TPP 429
>CO983154
Length = 568
Score = 85.1 bits (209), Expect = 8e-17
Identities = 51/136 (37%), Positives = 81/136 (59%), Gaps = 6/136 (4%)
Frame = +3
Query: 472 PQQNGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPED 531
PQQNG+AER R LLE AR ++ N + W +A T+C L+NR P S+L+ ++P
Sbjct: 9 PQQNGIAERKNRHLLETARSLMLNLNV--PIHHWGDAVLTSCFLINRMPSSSLENQIPHS 182
Query: 532 -IWSGNLVDYSNLRIFGCPAYALVND-----GKLAPRAVECIFLSYASESKGYRLWCSDP 585
++ + + + + ++FGC + V+D KL+ R+V+C+FL Y+ KGY+ C P
Sbjct: 183 LVFPHDPLFHVSPKVFGCTCF--VHDLSPGLDKLSARSVKCVFLGYSRLQKGYK--CYSP 350
Query: 586 KSQKLILSRDVTFNED 601
++ +S DVTF ED
Sbjct: 351 TMRRYYMSADVTFFED 398
>TC235104 weakly similar to UP|Q850H8 (Q850H8) Gag-pol polyprotein
(Fragment), partial (28%)
Length = 865
Score = 84.3 bits (207), Expect = 1e-16
Identities = 61/183 (33%), Positives = 94/183 (51%), Gaps = 2/183 (1%)
Frame = +2
Query: 316 KLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLE--FCKHYVFWKQKKVSFSTATHRTKGI 373
KL H LGH LSK ++ ++K++ FC+ K + S R
Sbjct: 308 KLLHERLGHPH------LSKL-KIMVPSLEKIKDLFCESCQLGKHVRSSXRHVESRVDSP 466
Query: 374 LDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKWRILVETQT 433
IH D+WGP++V+S RY +T ID+F + V+ ++ ++E +F ++TQ
Sbjct: 467 FLVIHXDIWGPNRVSSMS-YRYFVTFIDEFSQCTRVFLMKERSEIL-SFLTSVNKIKTQF 640
Query: 434 GKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCML 493
GK +K L +DN E+ SS + F + GI + P PQQN +AER R L+E AR +L
Sbjct: 641 GKTIKILRSDNAKEYFSSVISPFXSAQGILHQFSCPHTPQQNDIAERKNRHLVETARTLL 820
Query: 494 SNA 496
+A
Sbjct: 821 LHA 829
>TC222001 similar to UP|C716_NEPRA (O04164) Cytochrome P450 71A6 (Fragment)
, partial (21%)
Length = 912
Score = 64.3 bits (155), Expect = 1e-10
Identities = 42/106 (39%), Positives = 56/106 (52%), Gaps = 5/106 (4%)
Frame = -2
Query: 503 DLWVEAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYA---LVNDGKL 559
+ W A A +L+N P L P + G++ D S+LRIFGC YA N KL
Sbjct: 902 NFWNYALLHAAYLINCIPTPFLQNTSPYERLHGHIPDISHLRIFGCLCYASTIKANRKKL 723
Query: 560 APRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTF--NEDAL 603
PRA CIF+ + +KGY L+ D S +I SR+V F N DA+
Sbjct: 722 EPRAHPCIFIGFKPNTKGYMLY--DLHSHNIITSRNVVFYENHDAM 591
>BI425121
Length = 412
Score = 63.9 bits (154), Expect = 2e-10
Identities = 46/153 (30%), Positives = 67/153 (43%), Gaps = 6/153 (3%)
Frame = +3
Query: 389 SYGGRRYMMTIIDDFPRKVWVYFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN*LEF 448
S G ++Y +DD+ R WVYFL +K+E+ F + G +KK+ LE
Sbjct: 6 SLGCKKYEFLTVDDYSRYTWVYFLAHKHESLRYFIR---------GFKMKKVFVFLLLEV 158
Query: 449 CSS------DFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCMLSNAGL*N*R 502
N FC +GI + + PR PQ+N V ER RTL E+AR +L
Sbjct: 159 TMELSLRILS*NHFCERNGIFHNLS*PRTPQENRVVERKNRTLQEKARTIL--------- 311
Query: 503 DLWVEAASTACHLVNRSPHSALDFKVPEDIWSG 535
T C + N+ + K P ++W G
Sbjct: 312 -------FTTCFVQNKILIRPMIKKTPYELWKG 389
>CO981879
Length = 576
Score = 63.9 bits (154), Expect = 2e-10
Identities = 36/91 (39%), Positives = 51/91 (55%), Gaps = 8/91 (8%)
Frame = -2
Query: 518 RSPHSALDFKVPEDIWSG-----NLVDYSNLRIFGCPAYALV---NDGKLAPRAVECIFL 569
R P L+F+ P D+++ L L+IFGC + + N GKL PRA +C+F+
Sbjct: 284 RMPSKILNFRTPLDVFTSAFPNNRLSCTLPLKIFGCTVFVHIHEPNQGKLEPRAKKCVFV 105
Query: 570 SYASESKGYRLWCSDPKSQKLILSRDVTFNE 600
YA KGY+ C DP S+K ++ DVTF E
Sbjct: 104 GYAPNQKGYK--CFDPTSKKTFVTIDVTFFE 18
Score = 48.1 bits (113), Expect = 1e-05
Identities = 33/106 (31%), Positives = 52/106 (48%)
Frame = -1
Query: 422 FKKWRILVETQTGKNVKKLITDN*LEFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERM 481
FK + +++TQ +K +DN E+ + ++ +GI + PQQNGVAER
Sbjct: 567 FKTFFQMIQTQFQVKIKVFRSDNGREYFNKHLSKXXLENGIIHQSSCVDTPQQNGVAERK 388
Query: 482 IRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFK 527
R L E AR +L + W EA T +L N++ L+F+
Sbjct: 387 NRHLXEVARALLFQNKA--PKYXWGEAILTGTYLKNKNA*QNLEFQ 256
>BM892610
Length = 421
Score = 63.5 bits (153), Expect = 3e-10
Identities = 38/90 (42%), Positives = 56/90 (62%), Gaps = 3/90 (3%)
Frame = -3
Query: 514 HLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYALV---NDGKLAPRAVECIFLS 570
+L+NR P ++L F+VP +++ L +Y+ LR FGC Y + N KL R+ EC+FL
Sbjct: 413 YLINRLPTTSLKFQVPCTVFN-KLPNYNFLRNFGCSCYPFLRPYNKHKLDFRSHECLFLG 237
Query: 571 YASESKGYRLWCSDPKSQKLILSRDVTFNE 600
Y++ KGY+ C P KL +S+DV FNE
Sbjct: 236 YSTPHKGYK--CLSPNG-KLYVSKDVIFNE 156
>BI702130 weakly similar to GP|4234854|gb| pol polyprotein {Zea mays},
partial (14%)
Length = 421
Score = 51.6 bits (122), Expect(2) = 1e-09
Identities = 29/74 (39%), Positives = 45/74 (60%)
Frame = -1
Query: 477 VAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACHLVNRSPHSALDFKVPEDIWSGN 536
VAER RTLL+ R M SN L + LW++A TA +++NR P A+ K P +++ G
Sbjct: 250 VAERRNRTLLDMVRSMRSNVKL--PQFLWIDALKTAAYILNRVPTKAVS-KTPFELFKGW 80
Query: 537 LVDYSNLRIFGCPA 550
++R++GCP+
Sbjct: 79 KPSLRHIRVWGCPS 38
Score = 29.3 bits (64), Expect(2) = 1e-09
Identities = 18/57 (31%), Positives = 25/57 (43%), Gaps = 9/57 (15%)
Frame = -2
Query: 429 VETQTGKNVKKLITDN*LEF---------CSSDFNEFCTNHGIARHKTIPRNPQQNG 476
VE Q GK +K + +D E+ F +F H I T+P +P QNG
Sbjct: 420 VEKQCGKQIKIVRSDRGGEYYGRYTEDGQAPGSFAKFLQEHEIVAQYTMPGSPDQNG 250
>CO982898
Length = 816
Score = 46.6 bits (109), Expect(2) = 1e-08
Identities = 29/84 (34%), Positives = 45/84 (53%), Gaps = 3/84 (3%)
Frame = -1
Query: 520 PHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYALV---NDGKLAPRAVECIFLSYASESK 576
P +++F VP ++DYS R+FGC + L+ N K ++ ECIFL + K
Sbjct: 597 PTVSINFAVPYTQLFNQMLDYSFPRVFGCACFPLIRPYNIQKFNFKSHECIFLGNYTSHK 418
Query: 577 GYRLWCSDPKSQKLILSRDVTFNE 600
Y+ C P + ++S+DV FNE
Sbjct: 417 NYK--CLAPAGK--LVSKDVIFNE 358
Score = 31.2 bits (69), Expect(2) = 1e-08
Identities = 24/67 (35%), Positives = 32/67 (46%)
Frame = -3
Query: 455 EFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCMLSNAGL*N*RDLWVEAASTACH 514
EF + GI HK I + Q V ER R +++ +LS+A L W A T
Sbjct: 784 EFLND*GII-HKLIFPHTQYQNVVERKQRHIVKFGLSLLSHASLP--LKFWGHAFITPVF 614
Query: 515 LVNRSPH 521
L+NR PH
Sbjct: 613 LINRLPH 593
>CO986013
Length = 803
Score = 40.4 bits (93), Expect(2) = 9e-07
Identities = 24/67 (35%), Positives = 35/67 (51%), Gaps = 4/67 (5%)
Frame = -2
Query: 514 HLVNRSPHSALDFKVPEDI-WSGNLVDYSNLRIFG---CPAYALVNDGKLAPRAVECIFL 569
HL+NR S++D VP + + L+ + R+FG C Y KL RA++CIFL
Sbjct: 562 HLINRMASSSIDNNVPHYVLFLDELLSHIPPRVFGSICCVHYLTRGLDKLIARAIKCIFL 383
Query: 570 SYASESK 576
Y+ K
Sbjct: 382 GYSRNKK 362
Score = 30.8 bits (68), Expect(2) = 9e-07
Identities = 13/29 (44%), Positives = 18/29 (61%)
Frame = -1
Query: 573 SESKGYRLWCSDPKSQKLILSRDVTFNED 601
++ KGY WC P + + +S DVTF ED
Sbjct: 374 TQQKGY--WCYSPANHRFYMSADVTFFED 294
>BI788167
Length = 421
Score = 45.8 bits (107), Expect = 5e-05
Identities = 28/91 (30%), Positives = 43/91 (46%), Gaps = 17/91 (18%)
Frame = +2
Query: 209 FHMCPYRDLFTTFEPVDCGVVLMGNDVQCKVA*IGIVHFKTNDGVVRTLTN--------- 259
+HM P + F T+E + G V MGND ++ +G + K DG+VRT+
Sbjct: 71 WHMTPC*E*FCTYETILEGFVFMGNDHALEIVGVGTIKLKMYDGIVRTIQGVLHVKGLKK 250
Query: 260 --------ESLVCKYSAEGGVLMVSKGYLVL 282
+ L CK EG +LM S Y+++
Sbjct: 251 NQLFVGKLDDLECKIHIEGEILMGSFMYVMI 343
>BI425191 weakly similar to GP|14586969|gb| pol polyprotein {Citrus x
paradisi}, partial (7%)
Length = 423
Score = 45.8 bits (107), Expect = 5e-05
Identities = 27/86 (31%), Positives = 45/86 (51%)
Frame = +3
Query: 358 QKKVSFSTATHRTKGILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFLRYKNE 417
++K + AT T+ +L+ +H+D+ P V S+G RY +T IDD+ +VY L K +
Sbjct: 57 KEKHTMKRATRSTQ-LLEIMHTDICRPFDVNSFGKERYFITFIDDYSHYGYVYLLHEKFQ 233
Query: 418 TFPTFKKWRILVETQTGKNVKKLITD 443
+ VE Q + VK + +D
Sbjct: 234 AVDVLEIHLNEVERQLDRKVKVVRSD 311
>BI424202
Length = 421
Score = 45.1 bits (105), Expect = 9e-05
Identities = 28/101 (27%), Positives = 50/101 (48%)
Frame = -3
Query: 312 KDVTKLWHMHLGHMTEKGMHLLSKQGRLGKQGIDKLEFCKHYVFWKQKKVSFSTATHRTK 371
++ + LWH LGH++ + + L +G L E + KQ S A R+
Sbjct: 392 EESSMLWHRRLGHISIERIKRLVNEGVLSTLDFADFETYVDCIKGKQTNKSKKGAK-RSS 216
Query: 372 GILDYIHSDLWGPSKVTSYGGRRYMMTIIDDFPRKVWVYFL 412
+L+ IH+D+ P + +Y +T IDD+ R +++YF+
Sbjct: 215 NLLEIIHTDICCPDMDAN--SLKYFITFIDDYSRYMYLYFI 99
>TC223332
Length = 427
Score = 43.9 bits (102), Expect = 2e-04
Identities = 21/44 (47%), Positives = 27/44 (60%)
Frame = +2
Query: 558 KLAPRAVECIFLSYASESKGYRLWCSDPKSQKLILSRDVTFNED 601
KL RA++CIFL Y+ KGY WC + + +S DVTF ED
Sbjct: 29 KLKARAIKCIFLDYSCTQKGY--WCYSLANHRFYMSVDVTFFED 154
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.331 0.144 0.458
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,389,102
Number of Sequences: 63676
Number of extensions: 467021
Number of successful extensions: 3120
Number of sequences better than 10.0: 76
Number of HSP's better than 10.0 without gapping: 3067
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3096
length of query: 610
length of database: 12,639,632
effective HSP length: 103
effective length of query: 507
effective length of database: 6,081,004
effective search space: 3083069028
effective search space used: 3083069028
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC149197.1


