

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149130.7 - phase: 0 /pseudo
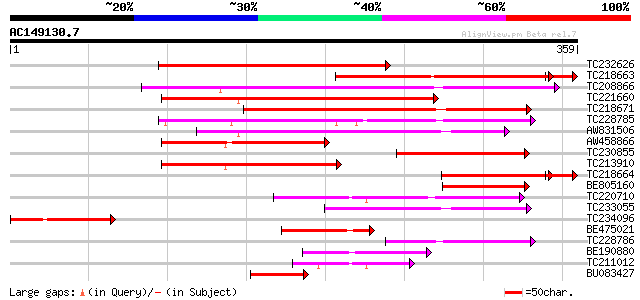
(359 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC232626 similar to GB|AAP37831.1|30725618|BT008472 At4g31350 {A... 223 1e-58
TC218663 weakly similar to GB|AAP37831.1|30725618|BT008472 At4g3... 202 6e-55
TC208866 weakly similar to GB|AAP68292.1|31711872|BT008853 At5g1... 209 2e-54
TC221660 similar to UP|Q9SY70 (Q9SY70) F14N23.16, partial (42%) 174 6e-44
TC218671 weakly similar to GB|AAP68292.1|31711872|BT008853 At5g1... 163 1e-40
TC228785 weakly similar to GB|AAP42749.1|30984572|BT008736 At3g5... 141 5e-34
AW831506 weakly similar to GP|4914330|gb| F14N23.16 {Arabidopsis... 139 2e-33
AW458866 117 1e-26
TC230855 similar to GB|AAP37842.1|30725640|BT008483 At5g57270 {A... 116 1e-26
TC213910 weakly similar to UP|Q75M40 (Q75M40) Unknow protein, pa... 111 6e-25
TC218664 weakly similar to GB|AAP37831.1|30725618|BT008472 At4g3... 100 2e-24
BE805160 97 1e-20
TC220710 similar to UP|Q8MLW3 (Q8MLW3) CG30386-PA, partial (7%) 93 2e-19
TC233055 weakly similar to UP|Q9SY70 (Q9SY70) F14N23.16, partial... 85 4e-17
TC234096 83 2e-16
BE475021 similar to GP|22535532|dbj P0431H09.21 {Oryza sativa (j... 74 1e-13
TC228786 weakly similar to GB|AAP42749.1|30984572|BT008736 At3g5... 64 1e-10
BE190880 51 9e-07
TC211012 similar to GB|AAP42749.1|30984572|BT008736 At3g52060 {A... 51 9e-07
BU083427 47 2e-05
>TC232626 similar to GB|AAP37831.1|30725618|BT008472 At4g31350 {Arabidopsis
thaliana;} , partial (44%)
Length = 888
Score = 223 bits (567), Expect = 1e-58
Identities = 100/147 (68%), Positives = 125/147 (85%)
Frame = +2
Query: 95 QNPKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKAKPVHVSRYFVNRDIRSDQL 154
+ PK+AF+FL+PGSLPFEKLW FFQGHEGKFSVYVH+SK KP HVS +FV R+I S+ +
Sbjct: 125 KKPKVAFLFLSPGSLPFEKLWHMFFQGHEGKFSVYVHSSKEKPTHVSSFFVGREIHSEPV 304
Query: 155 VWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRD 214
WGK+S+ EAERRLLA+AL DP+NQHFVLLS+SC+P+ F ++++YL+ T+ SF+DS+ D
Sbjct: 305 GWGKISMAEAERRLLAHALLDPDNQHFVLLSESCIPVRRFEFVYNYLLLTNVSFIDSYVD 484
Query: 215 PGPVGNGRYSEHMLPEVEIKDFRTGAQ 241
PGP GNGRY EHMLPEVE KDFR G+Q
Sbjct: 485 PGPHGNGRYIEHMLPEVEKKDFRKGSQ 565
>TC218663 weakly similar to GB|AAP37831.1|30725618|BT008472 At4g31350
{Arabidopsis thaliana;} , partial (47%)
Length = 915
Score = 202 bits (513), Expect(2) = 6e-55
Identities = 94/138 (68%), Positives = 113/138 (81%)
Frame = +2
Query: 207 SFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQ 266
SFVD F+DPGP GNGRYS+HMLPEVE+KDFR GAQWF++KRQHA+ VMAD+LYYSKF++
Sbjct: 2 SFVDCFKDPGPHGNGRYSDHMLPEVEVKDFRKGAQWFAMKRQHAIIVMADNLYYSKFRSY 181
Query: 267 CEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELL 326
C Q ++GKNCI DEHYLPTFF +VDP GIA WS+T+VD SE+K HPKSYR QD+TYELL
Sbjct: 182 C-QPGLEGKNCIADEHYLPTFFQMVDPGGIANWSLTHVDWSERKWHPKSYRAQDVTYELL 358
Query: 327 KNIKLHNFHLFLTERSTK 344
KNI + + +T K
Sbjct: 359 KNITSIDVSMHVTSDEKK 412
Score = 30.4 bits (67), Expect(2) = 6e-55
Identities = 13/20 (65%), Positives = 16/20 (80%)
Frame = +1
Query: 340 ERSTKMDLFLEWISEAVLPI 359
ERS K+ L +EW SEA+LPI
Sbjct: 409 ERSAKLALLMEWDSEAMLPI 468
>TC208866 weakly similar to GB|AAP68292.1|31711872|BT008853 At5g11730
{Arabidopsis thaliana;} , partial (78%)
Length = 1094
Score = 209 bits (532), Expect = 2e-54
Identities = 109/267 (40%), Positives = 156/267 (57%), Gaps = 2/267 (0%)
Frame = +2
Query: 84 YIERGITLDMPQNPKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHA--SKAKPVHVS 141
++ R + + PKIAFMFLT G LP LW+ FF+GHEG +S+YVH+ S S
Sbjct: 68 FVPRIKSYPFKRTPKIAFMFLTKGPLPMAPLWEKFFRGHEGLYSIYVHSLPSYNADFSPS 247
Query: 142 RYFVNRDIRSDQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYL 201
F R I S WG MS+ +AERRLLANAL D +N+ F+LLS+SC+PL NF+ ++ Y+
Sbjct: 248 SVFYRRQIPSQVAEWGMMSMCDAERRLLANALLDISNEWFILLSESCIPLQNFSIVYLYI 427
Query: 202 MYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYS 261
+ SF+ + +PGP G GRY +M PE+ + D+R G+QWF + R+ A++++ D+ YY
Sbjct: 428 ARSRYSFMGAVDEPGPYGRGRYDGNMAPEINMSDWRKGSQWFEINRELALRIVEDNTYYP 607
Query: 262 KFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDI 321
K + C+ C +DEHY T T P+ +A S+TYVD S HP ++ DI
Sbjct: 608 KLKEFCKP-----HKCYVDEHYFQTMLTSNTPHLLANRSLTYVDWSRGGAHPATFGKDDI 772
Query: 322 TYELLKNIKLHNFHLFLTERSTKMDLF 348
E K I L+ + S+ LF
Sbjct: 773 KEEFFKKILQDQTCLYNNQPSSLCFLF 853
>TC221660 similar to UP|Q9SY70 (Q9SY70) F14N23.16, partial (42%)
Length = 545
Score = 174 bits (441), Expect = 6e-44
Identities = 82/177 (46%), Positives = 119/177 (66%), Gaps = 2/177 (1%)
Frame = +2
Query: 97 PKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKAKPVHVSRY--FVNRDIRSDQL 154
PK+AFMFLT G LP LW+ FF GH F++Y+H+ ++VS F R I S +
Sbjct: 2 PKVAFMFLTRGPLPMLPLWERFFHGHSSLFNIYIHSPPRFLLNVSHSSPFYLRHIPSQDV 181
Query: 155 VWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRD 214
WG +++ +AERRLLANAL D +N+ FVLLS+SC+P+YNF ++ YL + SFV+S+ +
Sbjct: 182 SWGTVTLADAERRLLANALLDFSNERFVLLSESCIPVYNFPTVYRYLTNSSLSFVESYDE 361
Query: 215 PGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSC 271
P G GRYS +MLP ++++ +R G+QWF L R AV +++D YYS F+ C+ +C
Sbjct: 362 PTRYGRGRYSRNMLPHIQLRHWRKGSQWFELNRALAVYIVSDTNYYSLFRKYCKPAC 532
>TC218671 weakly similar to GB|AAP68292.1|31711872|BT008853 At5g11730
{Arabidopsis thaliana;} , partial (46%)
Length = 899
Score = 163 bits (412), Expect = 1e-40
Identities = 80/182 (43%), Positives = 121/182 (65%)
Frame = +2
Query: 149 IRSDQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSF 208
I S ++ WG ++++EAERRLLANAL D +NQ FVLLS+SC+PL+NF+ I+ YLM + +++
Sbjct: 8 IPSXEVEWGNVNMIEAERRLLANALVDISNQRFVLLSESCIPLFNFSTIYTYLMNSTQNY 187
Query: 209 VDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCE 268
V + DP VG GRYS MLPE+ + +R G+QWF + R A++V++D Y+ FQ C+
Sbjct: 188 VMAVDDPSSVGRGRYSIQMLPEISLNQWRKGSQWFEMDRDLALEVVSDRKYFPVFQDYCK 367
Query: 269 QSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKN 328
S C DEHYLPT+ +I G + S+T+VD S+ HP + +IT + L++
Sbjct: 368 GS------CYADEHYLPTYVSIKFWEGNSNRSLTWVDWSKGGPHPTKFLRSEITVKFLES 529
Query: 329 IK 330
++
Sbjct: 530 LR 535
>TC228785 weakly similar to GB|AAP42749.1|30984572|BT008736 At3g52060
{Arabidopsis thaliana;} , partial (65%)
Length = 860
Score = 141 bits (355), Expect = 5e-34
Identities = 89/272 (32%), Positives = 141/272 (51%), Gaps = 33/272 (12%)
Frame = +3
Query: 95 QNP--KIAFMFLTPGSLPFEKLWDNFFQGHEGK-FSVYVHASKAKPVH--VSRYFVNRDI 149
+NP KIAF+FLT L F LWD FF F++Y+HA + + +S F+N+ I
Sbjct: 39 KNPSLKIAFLFLTNSDLHFSSLWDQFFSNTPSNLFNIYIHADPSVNLTRPLSPLFINKFI 218
Query: 150 RSDQLVWGKMSIVEAERRLLANAL-QDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTD--- 205
S + +++ A RRLLA AL DP+N +F LLS C+PL++F+Y + L +
Sbjct: 219 SSKRTFRSSPTLISATRRLLATALLDDPSNAYFALLSQHCIPLHSFSYTYKSLFLSPTFD 398
Query: 206 ------------------KSFVDSFRDPGPV-----GNGRYSEHMLPEVEIKDFRTGAQW 242
KSFV+ + GRY+ M+PE+ + FR G+Q+
Sbjct: 399 SQDPESSSSTRFGLRLKYKSFVEILSHAPKLWKRYTSRGRYA--MMPEIPFEAFRVGSQF 572
Query: 243 FSLKRQHAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVT 302
F+L R+HA+ V+ D + KF+ C + C +EHY PT ++ DP+G K+++T
Sbjct: 573 FTLTRRHALVVVKDRTLWQKFKIPCYRD----DECYPEEHYFPTLLSMADPDGCTKYTLT 740
Query: 303 YVD-RSEQKRHPKSYRTQDITYELLKNIKLHN 333
V+ HP +YR +++ EL+ ++ N
Sbjct: 741 RVNWTGTVNGHPYTYRPTEVSPELILRLRKSN 836
>AW831506 weakly similar to GP|4914330|gb| F14N23.16 {Arabidopsis thaliana},
partial (44%)
Length = 589
Score = 139 bits (350), Expect = 2e-33
Identities = 73/200 (36%), Positives = 115/200 (57%), Gaps = 2/200 (1%)
Frame = +2
Query: 119 FQGHEGKFSVYVHASKAKPVHVSRY--FVNRDIRSDQLVWGKMSIVEAERRLLANALQDP 176
F GH FS+Y+HA +++S F R+I S + WG ++ +A RRLLA+AL
Sbjct: 2 FHGHSSLFSIYIHAPPRCTLNISHSSPFYLRNIPSQDVSWGTFTLADAHRRLLADALLHF 181
Query: 177 NNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEIKDF 236
+N+ F+LLS++C+P Y+ + L + S V+S+ +P G GRYS HML + ++ +
Sbjct: 182 SNERFLLLSETCIPGYDLPTAYTDLTHYYLSCVESYDEPTRYGRGRYSRHMLAHIHLRHW 361
Query: 237 RTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGI 296
R G+QW L R AV +++D YYS F+ C+ +C DEHY+PTF + +
Sbjct: 362 RKGSQWLELNRSLAVYIVSDTKYYSLFRKYCKPACYP------DEHYIPTFLHMFHGSLN 523
Query: 297 AKWSVTYVDRSEQKRHPKSY 316
+ +VT+VD S HP ++
Sbjct: 524 SNRTVTWVDWSMLGAHPATF 583
>AW458866
Length = 388
Score = 117 bits (292), Expect = 1e-26
Identities = 58/110 (52%), Positives = 80/110 (72%), Gaps = 4/110 (3%)
Frame = +2
Query: 97 PKIAFMFLTPGSLPFEKLWDNFFQGHEGKFSVYVHASKA----KPVHVSRYFVNRDIRSD 152
PK+AFMFLT G + LW+ FF+GHEG +S+YVH++ + +P S F R I S
Sbjct: 50 PKVAFMFLTRGPVFLAPLWEQFFKGHEGFYSIYVHSNPSYNGSRPE--SPVFKGRRIPSK 223
Query: 153 QLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLM 202
++ WG ++++EAERRLLANAL D +NQ FVLLS+SC+PL+NF+ I+ YLM
Sbjct: 224 EVEWGNVNMIEAERRLLANALVDISNQRFVLLSESCIPLFNFSTIYTYLM 373
>TC230855 similar to GB|AAP37842.1|30725640|BT008483 At5g57270 {Arabidopsis
thaliana;} , partial (24%)
Length = 418
Score = 116 bits (291), Expect = 1e-26
Identities = 50/84 (59%), Positives = 66/84 (78%)
Frame = +3
Query: 246 KRQHAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVD 305
KRQHA+ VMAD LY++KF+ C + +NC DEHYLPTFF ++DP GIA WSVTYVD
Sbjct: 3 KRQHAIIVMADSLYFTKFKHHCRPNMEGNRNCYADEHYLPTFFIMLDPGGIANWSVTYVD 182
Query: 306 RSEQKRHPKSYRTQDITYELLKNI 329
S++K HP+S+R +DITY+++KNI
Sbjct: 183 WSQRKWHPRSFRARDITYQVMKNI 254
>TC213910 weakly similar to UP|Q75M40 (Q75M40) Unknow protein, partial (29%)
Length = 589
Score = 111 bits (277), Expect = 6e-25
Identities = 59/120 (49%), Positives = 82/120 (68%), Gaps = 6/120 (5%)
Frame = +1
Query: 97 PKIAFMFLTPGSLPFEKLWDNFFQ-GHEGKFSVYVHASKA----KPVHVSRYFVNRDI-R 150
P+ +FL +LP + LWD FFQ G +FS+YVH++ + S+ F R I
Sbjct: 193 PRSLSLFLVRRNLPLDFLWDAFFQNGDVSRFSIYVHSAPGFVLDESTTRSQLFYGRQISN 372
Query: 151 SDQLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVD 210
S Q++WG+ S+++AER LLA AL+D NQ FVLLSDSCVPLYNF+Y+++YLM + +SFVD
Sbjct: 373 SIQVLWGESSMIQAERLLLAAALEDHANQRFVLLSDSCVPLYNFSYVYNYLMASPRSFVD 552
>TC218664 weakly similar to GB|AAP37831.1|30725618|BT008472 At4g31350
{Arabidopsis thaliana;} , partial (29%)
Length = 673
Score = 99.8 bits (247), Expect(2) = 2e-24
Identities = 46/71 (64%), Positives = 54/71 (75%)
Frame = +3
Query: 274 GKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKNIKLHN 333
GKNCI DEHYLPTFF +VDP GIA WS+T+VD SE+K HPKSYR QD+TYELLKNI +
Sbjct: 3 GKNCIXDEHYLPTFFQMVDPGGIANWSLTHVDWSERKWHPKSYRAQDVTYELLKNITSID 182
Query: 334 FHLFLTERSTK 344
+ +T K
Sbjct: 183 VSVHVTSDEKK 215
Score = 30.4 bits (67), Expect(2) = 2e-24
Identities = 13/20 (65%), Positives = 16/20 (80%)
Frame = +2
Query: 340 ERSTKMDLFLEWISEAVLPI 359
ERS K+ L +EW SEA+LPI
Sbjct: 212 ERSAKLALLMEWDSEAMLPI 271
>BE805160
Length = 351
Score = 97.1 bits (240), Expect = 1e-20
Identities = 43/55 (78%), Positives = 48/55 (87%)
Frame = +2
Query: 275 KNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQDITYELLKNI 329
KNCI DEHYLPTFF +VDP GIA WS+T+VD SE+K HPKSYR QD+TYELLKNI
Sbjct: 2 KNCIADEHYLPTFFQMVDPGGIANWSLTHVDWSERKWHPKSYRAQDVTYELLKNI 166
>TC220710 similar to UP|Q8MLW3 (Q8MLW3) CG30386-PA, partial (7%)
Length = 496
Score = 92.8 bits (229), Expect = 2e-19
Identities = 59/165 (35%), Positives = 91/165 (54%), Gaps = 6/165 (3%)
Frame = +1
Query: 168 LLANALQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTD-KSFVDSFRDPGPVGNGRYS-- 224
L A L DP N +F LLS C+PL++ + ++L KSF++ + P RY+
Sbjct: 7 LAAALLDDPLNHYFALLSQYCIPLHSLQFTHNFLFKNPHKSFIEILSNE-PNLFDRYTAR 183
Query: 225 -EH-MLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEH 282
EH MLPE+ FR G+Q+F L R+HA V+ D L ++KF+ CV + C +EH
Sbjct: 184 GEHAMLPEIPFSSFRVGSQFFILTRRHARVVVRDILLWNKFRL----PCVTEEPCYPEEH 351
Query: 283 YLPTFFTIVDPNGIAKWSVTYVDRSE-QKRHPKSYRTQDITYELL 326
Y PT ++ DPNG +++T V+ + HP Y +++ EL+
Sbjct: 352 YFPTLLSMQDPNGCTGFTLTRVNWTGCWDGHPHLYTAPEVSPELI 486
>TC233055 weakly similar to UP|Q9SY70 (Q9SY70) F14N23.16, partial (38%)
Length = 718
Score = 85.1 bits (209), Expect = 4e-17
Identities = 44/131 (33%), Positives = 70/131 (52%)
Frame = +2
Query: 200 YLMYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSLKRQHAVKVMADHLY 259
Y+ ++ S +S+ DP G G S H P + + +R G QWF L R AV +++D Y
Sbjct: 14 YVTHSSISXXESYDDPARYGRGXXSRHXXPHIXXRHWRKGXQWFELNRSLAVYIVSDTKY 193
Query: 260 YSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAKWSVTYVDRSEQKRHPKSYRTQ 319
YS F+ C+ +C DEHY+PTF + + + +VT+VD S HP ++
Sbjct: 194 YSLFRKYCKPACYP------DEHYIPTFLHMFHGSLNSNRTVTWVDWSMLGPHPATFGRA 355
Query: 320 DITYELLKNIK 330
+IT L++I+
Sbjct: 356 NITAAFLQSIR 388
>TC234096
Length = 435
Score = 82.8 bits (203), Expect = 2e-16
Identities = 41/68 (60%), Positives = 50/68 (73%), Gaps = 1/68 (1%)
Frame = +3
Query: 1 MKTAKFWRLS-MGDMEILLGSPVLHRPQMKKPMWIVVLVSFIILFLTCAYLYRMQNTTSC 59
MKT K WRL MGDM+IL GS HRP MKKP WI+VLV F+ +FL CAY+Y QN+++C
Sbjct: 225 MKTEKVWRLGGMGDMQILPGSR--HRPPMKKPKWIIVLVLFVCVFLICAYIYPPQNSSTC 398
Query: 60 NMFYSKPC 67
+F SK C
Sbjct: 399 YVFSSKGC 422
>BE475021 similar to GP|22535532|dbj P0431H09.21 {Oryza sativa (japonica
cultivar-group)}, partial (17%)
Length = 178
Score = 73.9 bits (180), Expect = 1e-13
Identities = 35/59 (59%), Positives = 44/59 (74%)
Frame = +1
Query: 173 LQDPNNQHFVLLSDSCVPLYNFNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPEV 231
L+DP NQ FVLLSDSCVPLYNF+Y+++YLM + KSFVDSF D G Y+ M P++
Sbjct: 1 LEDPANQRFVLLSDSCVPLYNFSYMYNYLMVSPKSFVDSFLD---AKEGHYNPKMSPKI 168
>TC228786 weakly similar to GB|AAP42749.1|30984572|BT008736 At3g52060
{Arabidopsis thaliana;} , partial (36%)
Length = 826
Score = 63.9 bits (154), Expect = 1e-10
Identities = 32/96 (33%), Positives = 55/96 (56%), Gaps = 1/96 (1%)
Frame = +2
Query: 239 GAQWFSLKRQHAVKVMADHLYYSKFQAQCEQSCVDGKNCILDEHYLPTFFTIVDPNGIAK 298
G+Q+F+L R+HA+ V+ D + KF+ C + C +EHY PT ++ DP+G K
Sbjct: 5 GSQFFTLTRRHALVVVKDRTLWRKFKIPCYRD----DECYPEEHYFPTLLSMADPDGCTK 172
Query: 299 WSVTYVD-RSEQKRHPKSYRTQDITYELLKNIKLHN 333
+++T V+ HP +YR +I+ EL+ ++ N
Sbjct: 173 YTLTSVNWTGTVNGHPYTYRPTEISPELILRLRKSN 280
>BE190880
Length = 241
Score = 50.8 bits (120), Expect = 9e-07
Identities = 27/82 (32%), Positives = 42/82 (50%)
Frame = -1
Query: 186 DSCVPLYNFNYIFDYLMYTDKSFVDSFRDPGPVGNGRYSEHMLPEVEIKDFRTGAQWFSL 245
D+C+PLY + ++ +M + SFVDS D GRY+ +M P + + + R +QW L
Sbjct: 238 DTCIPLYTSSNTYESIMSSSTSFVDSLLDR---KTGRYNPYMDPVIPVYNRRIRSQWAVL 68
Query: 246 KRQHAVKVMADHLYYSKFQAQC 267
R+H V+ FQ C
Sbjct: 67 TRKHTKVVVQYETVVPMFQRYC 2
>TC211012 similar to GB|AAP42749.1|30984572|BT008736 At3g52060 {Arabidopsis
thaliana;} , partial (20%)
Length = 326
Score = 50.8 bits (120), Expect = 9e-07
Identities = 33/85 (38%), Positives = 48/85 (55%), Gaps = 8/85 (9%)
Frame = +1
Query: 180 HFVLLSDSCVPLYNF----NYIFDYLMYTDKSFVDSFRDPGPVGNGRYS---EH-MLPEV 231
+F LLS C+PL++ N++F + KSF++ + P RY+ EH MLPEV
Sbjct: 7 YFALLSQHCIPLHSLQFTHNFLFKNPTHPHKSFIEILSNE-PNLFDRYTARGEHAMLPEV 183
Query: 232 EIKDFRTGAQWFSLKRQHAVKVMAD 256
FR G+Q+F L R+HA V+ D
Sbjct: 184 PFSSFRVGSQFFILTRRHARTVVRD 258
>BU083427
Length = 340
Score = 46.6 bits (109), Expect = 2e-05
Identities = 22/37 (59%), Positives = 29/37 (77%)
Frame = -1
Query: 153 QLVWGKMSIVEAERRLLANALQDPNNQHFVLLSDSCV 189
Q++WG+ S+++AER LLA AL+D NQ FVLLSD V
Sbjct: 262 QVLWGESSMIQAERLLLAAALEDHANQRFVLLSDRLV 152
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.139 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,354,722
Number of Sequences: 63676
Number of extensions: 278237
Number of successful extensions: 1962
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 1931
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1940
length of query: 359
length of database: 12,639,632
effective HSP length: 98
effective length of query: 261
effective length of database: 6,399,384
effective search space: 1670239224
effective search space used: 1670239224
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC149130.7


