

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149130.3 - phase: 0 /pseudo
(171 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
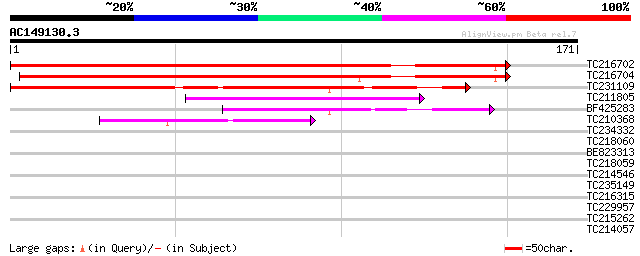
Score E
Sequences producing significant alignments: (bits) Value
TC216702 similar to UP|Q9LDP5 (Q9LDP5) FLORICAULA/LEAFY-like pro... 146 5e-36
TC216704 similar to UP|Q7PIN1 (Q7PIN1) ENSANGP00000023661 (Fragm... 127 3e-30
TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, par... 108 2e-24
TC211805 similar to UP|Q9K6E7 (Q9K6E7) BH3782 protein, partial (... 56 1e-08
BF425283 53 6e-08
TC210368 40 5e-04
TC234332 39 0.002
TC218060 35 0.018
BE823313 34 0.030
TC218059 weakly similar to UP|Q84KQ9 (Q84KQ9) F-box, partial (7%) 34 0.039
TC214546 similar to PIR|T01932|T01932 RNA binding protein homolo... 30 0.43
TC235149 30 0.43
TC216315 similar to GB|AAF36454.1|7108521|AF127564 ubiquitin-pro... 28 1.6
TC229957 weakly similar to UP|Q9LKB2 (Q9LKB2) Arabidopsis thalia... 28 1.6
TC215262 similar to UP|Q94C19 (Q94C19) At1g26850/T2P11_4, partia... 27 4.8
TC214057 homologue to UP|Q8GTE3 (Q8GTE3) Ribosomal protein S3a, ... 27 6.3
>TC216702 similar to UP|Q9LDP5 (Q9LDP5) FLORICAULA/LEAFY-like protein,
partial (5%)
Length = 1631
Score = 146 bits (368), Expect = 5e-36
Identities = 76/153 (49%), Positives = 104/153 (67%), Gaps = 2/153 (1%)
Frame = +3
Query: 1 MGVFVENSIRCIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGC 60
MGVFV NS+ ++ +KL+ P LI+AF+LT +IF E+PLP + ID+A+LGG
Sbjct: 303 MGVFVGNSLHWVVTRKLEPDQPDLIIAFDLTHDIFRELPLPDTGGVDGGFEIDLALLGGS 482
Query: 61 LCITVNYETTKIDVWVMKEYGYRDSWCKLFTLVKSCFTSHLRYSNPIGYSSDGSKVLFEG 120
LC+TVN+ T+IDVWVM+EY RDSWCK+FTL +S L+ P+GYSSDG+K
Sbjct: 483 LCMTVNFHKTRIDVWVMREYNRRDSWCKVFTLEESREMRSLKCVRPLGYSSDGNK----- 647
Query: 121 IEVLLDVHYKKLFWYDLKSERVSYV--EGIPKL 151
VLL+ K+LFWYDL+ + V+ V +G+P L
Sbjct: 648 --VLLEHDRKRLFWYDLEKKEVALVKIQGLPNL 740
>TC216704 similar to UP|Q7PIN1 (Q7PIN1) ENSANGP00000023661 (Fragment),
partial (5%)
Length = 1232
Score = 127 bits (319), Expect = 3e-30
Identities = 72/151 (47%), Positives = 96/151 (62%), Gaps = 3/151 (1%)
Frame = +2
Query: 4 FVENSIRCIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCI 63
FV NS+ ++ +KL+ P LIVAF+LT EIF E+PLP IDVA+LG LC+
Sbjct: 2 FVGNSLHWVVTRKLEPDQPDLIVAFDLTHEIFTELPLPDTGGVGGGFEIDVALLGDSLCM 181
Query: 64 TVNYETTKIDVWVMKEYGYRDSWCKLFTLVKSCFTSHLRYS-NPIGYSSDGSKVLFEGIE 122
TVN+ +K+DVWVM+EY DSWCKLFTL +S ++ P+GYSSDG+K
Sbjct: 182 TVNFHNSKMDVWVMREYNRGDSWCKLFTLEESA*VEIVQXCLRPLGYSSDGNK------- 340
Query: 123 VLLDVHYKKLFWYDLKSERVSYV--EGIPKL 151
VLL+ K+L WYDL + V+ V +G+P L
Sbjct: 341 VLLEHDRKRLCWYDLGKKEVTLVRIQGLPNL 433
>TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, partial (8%)
Length = 1061
Score = 108 bits (269), Expect = 2e-24
Identities = 63/145 (43%), Positives = 89/145 (60%), Gaps = 6/145 (4%)
Frame = +3
Query: 1 MGVFVENSIRCIMAKKLDGLHPSLIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGC 60
MGVFV S+ ++ +KL P LIV+F+LT E F+EVPLP + G+ + VA+LGGC
Sbjct: 660 MGVFVSGSLHWLVTRKLQPHEPDLIVSFDLTRETFHEVPLPVTVNGDFDM--QVALLGGC 833
Query: 61 LCITVNYETTKIDVWVMKEYGYRDSWCKLFTLVKS------CFTSHLRYSNPIGYSSDGS 114
LC+ V + T DVWVM+ YG R+SW KLFTL+++ + L+Y P+ + DG
Sbjct: 834 LCV-VEHRGTGFDVWVMRVYGSRNSWEKLFTLLENNDHHEMMGSGKLKYVRPL--ALDGD 1004
Query: 115 KVLFEGIEVLLDVHYKKLFWYDLKS 139
+VLFE + KL WY+LK+
Sbjct: 1005RVLFEHNRI-------KLCWYNLKT 1058
>TC211805 similar to UP|Q9K6E7 (Q9K6E7) BH3782 protein, partial (13%)
Length = 693
Score = 55.8 bits (133), Expect = 1e-08
Identities = 26/72 (36%), Positives = 41/72 (56%)
Frame = +1
Query: 54 VAVLGGCLCITVNYETTKIDVWVMKEYGYRDSWCKLFTLVKSCFTSHLRYSNPIGYSSDG 113
+ VL GCLC+ +Y+ T VW+MK+YG R+SW KL ++ + YS P S +G
Sbjct: 4 LGVLQGCLCMNYDYKKTHFVVWMMKDYGARESWVKLVSIPYVPNPENFSYSGPYYISENG 183
Query: 114 SKVLFEGIEVLL 125
+L +++L
Sbjct: 184 EVLLMFEFDLIL 219
>BF425283
Length = 336
Score = 53.1 bits (126), Expect = 6e-08
Identities = 34/86 (39%), Positives = 46/86 (52%), Gaps = 4/86 (4%)
Frame = +1
Query: 65 VNYETTKIDVWVMKEYGYRDSWCKLFTLVKS----CFTSHLRYSNPIGYSSDGSKVLFEG 120
V + T VWVM+ YG RDSW KLF+L ++ + L+Y P+ DG +VLF
Sbjct: 7 VEHRGTGFHVWVMRVYGSRDSWEKLFSLTENHHHEMGSGKLKYVRPLAL-DDGDRVLFXH 183
Query: 121 IEVLLDVHYKKLFWYDLKSERVSYVE 146
+ KL WYDLK+ VS V+
Sbjct: 184 -------NRXKLCWYDLKTGDVSCVK 240
>TC210368
Length = 815
Score = 40.0 bits (92), Expect = 5e-04
Identities = 20/67 (29%), Positives = 34/67 (49%), Gaps = 2/67 (2%)
Frame = +1
Query: 28 FNLTLEIFNEVPLPAELQG--ETSLLIDVAVLGGCLCITVNYETTKIDVWVMKEYGYRDS 85
F+L +E+PLP + + + + ++GGCL + + ++WVMKEY R S
Sbjct: 1 FDLVERTLSEIPLPLADRSTVQKDAVYGLRIMGGCLSVCCS-SCGMTEIWVMKEYKVRSS 177
Query: 86 WCKLFTL 92
W F +
Sbjct: 178WTMTFLI 198
>TC234332
Length = 644
Score = 38.5 bits (88), Expect = 0.002
Identities = 18/69 (26%), Positives = 34/69 (49%)
Frame = +2
Query: 24 LIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITVNYETTKIDVWVMKEYGYR 83
+I +++L E + + +P L ++ VL GCLC++ + W+MKE+G
Sbjct: 110 VIFSYDLKNESYRYLLMPDGLLEVPHSPPELVVLKGCLCLSHRHGGNHFGFWLMKEFGVE 289
Query: 84 DSWCKLFTL 92
SW + +
Sbjct: 290 KSWTRFLNI 316
>TC218060
Length = 901
Score = 35.0 bits (79), Expect = 0.018
Identities = 20/79 (25%), Positives = 38/79 (47%), Gaps = 5/79 (6%)
Frame = +2
Query: 19 GLHPSLIVAFNLTLEIFNEVPLP-----AELQGETSLLIDVAVLGGCLCITVNYETTKID 73
G +++AF++ E F ++ +P ++ + T + + + G L V D
Sbjct: 56 GATQDIVLAFDMVKESFRKIRVPKVRDSSDEKFATLVPFEESASIGVLVYPVRGAEKSFD 235
Query: 74 VWVMKEYGYRDSWCKLFTL 92
VWVMK+Y SW K +++
Sbjct: 236 VWVMKDYWDEGSWVKQYSV 292
>BE823313
Length = 503
Score = 34.3 bits (77), Expect = 0.030
Identities = 23/71 (32%), Positives = 36/71 (50%), Gaps = 6/71 (8%)
Frame = -1
Query: 33 EIFNEVPLPAELQGETSLLIDVAVL--GGCLCITVNYETT----KIDVWVMKEYGYRDSW 86
E+F EV LP L + + + V V+ G +TV + + + VMKEYG +SW
Sbjct: 500 EMFGEVMLPXXLAHVSXVAVVVKVVXGGNGXTLTVYHVSACYPCSCEXXVMKEYGVVESW 321
Query: 87 CKLFTLVKSCF 97
K+F+ + F
Sbjct: 320 NKVFSFSMNAF 288
>TC218059 weakly similar to UP|Q84KQ9 (Q84KQ9) F-box, partial (7%)
Length = 1185
Score = 33.9 bits (76), Expect = 0.039
Identities = 19/74 (25%), Positives = 38/74 (50%), Gaps = 5/74 (6%)
Frame = +3
Query: 24 LIVAFNLTLEIFNEVPLP-----AELQGETSLLIDVAVLGGCLCITVNYETTKIDVWVMK 78
+++AF++ E F ++ +P ++ + T + + + G L V + DVWVMK
Sbjct: 777 VVLAFDMVKESFRKIRVPKIRDSSDEKFGTLVPFEESASIGFLVYPVRGTEKRFDVWVMK 956
Query: 79 EYGYRDSWCKLFTL 92
+Y SW K +++
Sbjct: 957 DYWDEGSWVKQYSV 998
>TC214546 similar to PIR|T01932|T01932 RNA binding protein homolog - common
tobacco (fragment) {Nicotiana tabacum;} , partial (68%)
Length = 1756
Score = 30.4 bits (67), Expect = 0.43
Identities = 13/54 (24%), Positives = 29/54 (53%)
Frame = -3
Query: 22 PSLIVAFNLTLEIFNEVPLPAELQGETSLLIDVAVLGGCLCITVNYETTKIDVW 75
P L++ NLTL + ++P+ L L+ +V++ C+C+ ++ + + W
Sbjct: 257 PLLLLLLNLTLLVIIQIPVGTGLSHVFLLIKNVSLTCVCVCVCSSFSFSLLKTW 96
>TC235149
Length = 435
Score = 30.4 bits (67), Expect = 0.43
Identities = 17/78 (21%), Positives = 36/78 (45%), Gaps = 5/78 (6%)
Frame = +2
Query: 74 VWVMKEYGYRDSWCKLFTLVKSCFTSH-----LRYSNPIGYSSDGSKVLFEGIEVLLDVH 128
VW+ +E+G SW +L + F +H R+ P+ S + E + +L +
Sbjct: 8 VWLTREFGVERSWTRLLNVSYEHFRNHGCPPYYRFVTPLCMSEN------EDVLLLANDE 169
Query: 129 YKKLFWYDLKSERVSYVE 146
+ +Y+L+ R+ ++
Sbjct: 170 GSEFVFYNLRDNRIDRIQ 223
>TC216315 similar to GB|AAF36454.1|7108521|AF127564 ubiquitin-protein ligase
1 {Arabidopsis thaliana;} , partial (12%)
Length = 1938
Score = 28.5 bits (62), Expect = 1.6
Identities = 20/58 (34%), Positives = 30/58 (51%), Gaps = 2/58 (3%)
Frame = +1
Query: 98 TSHLRYSNPIGYSSDGSKVLFEGIEVLLDVHYKKLFWYDLKSERVSY--VEGIPKLFF 153
T HL Y +G K LF+G LLDVH+ + F+ + +V+Y +E I +F
Sbjct: 919 TEHLSYFKFVGRVV--GKALFDG--QLLDVHFTRSFYKHVLGAKVTYHDIEAIDPDYF 1080
>TC229957 weakly similar to UP|Q9LKB2 (Q9LKB2) Arabidopsis thaliana genomic
DNA, chromosome 3, TAC clone:K15M2, partial (9%)
Length = 836
Score = 28.5 bits (62), Expect = 1.6
Identities = 19/60 (31%), Positives = 27/60 (44%)
Frame = +3
Query: 51 LIDVAVLGGCLCITVNYETTKIDVWVMKEYGYRDSWCKLFTLVKSCFTSHLRYSNPIGYS 110
++D+A+LG + T K D+ V KE + + L KSC S Y N YS
Sbjct: 552 VMDIAILGKLAAQITQFSTIKSDIRVTKEV----TLILVLILSKSCSFSRFPYKNRELYS 719
>TC215262 similar to UP|Q94C19 (Q94C19) At1g26850/T2P11_4, partial (97%)
Length = 2422
Score = 26.9 bits (58), Expect = 4.8
Identities = 7/20 (35%), Positives = 15/20 (75%)
Frame = -2
Query: 70 TKIDVWVMKEYGYRDSWCKL 89
T +++W +E G ++SWC++
Sbjct: 60 TGLEIWDSQERGVKESWCRI 1
>TC214057 homologue to UP|Q8GTE3 (Q8GTE3) Ribosomal protein S3a, complete
Length = 1044
Score = 26.6 bits (57), Expect = 6.3
Identities = 18/60 (30%), Positives = 31/60 (51%), Gaps = 1/60 (1%)
Frame = +1
Query: 92 LVKSCFTSHLRY-SNPIGYSSDGSKVLFEGIEVLLDVHYKKLFWYDLKSERVSYVEGIPK 150
+++SCF SH + + +G + SK G + D KK WYD+K+ + V+ + K
Sbjct: 25 IIRSCFYSHRKQ*AMAVGKNKRISKGKKGGKKKAADPFAKK-DWYDIKAPSLFQVKNVGK 201
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.143 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,578,743
Number of Sequences: 63676
Number of extensions: 121730
Number of successful extensions: 752
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 743
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 744
length of query: 171
length of database: 12,639,632
effective HSP length: 90
effective length of query: 81
effective length of database: 6,908,792
effective search space: 559612152
effective search space used: 559612152
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 55 (25.8 bits)
Medicago: description of AC149130.3


