

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149129.2 + phase: 0
(261 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
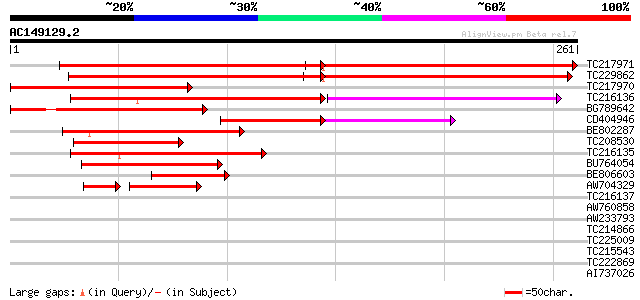
Score E
Sequences producing significant alignments: (bits) Value
TC217971 similar to UP|Q940L9 (Q940L9) At2g44770/F16B22.26, part... 229 e-107
TC229862 similar to UP|Q940L9 (Q940L9) At2g44770/F16B22.26, part... 205 2e-53
TC217970 155 1e-38
TC216136 121 4e-33
BG789642 122 1e-28
CD404946 83 1e-19
BE802287 weakly similar to GP|9828622|gb|A F1N21.22 {Arabidopsis... 87 1e-17
TC208530 80 9e-16
TC216135 75 3e-14
BU764054 weakly similar to GP|9828622|gb|A F1N21.22 {Arabidopsis... 75 3e-14
BE806603 68 5e-12
AW704329 weakly similar to GP|9828622|gb|A F1N21.22 {Arabidopsis... 35 1e-05
TC216137 35 0.026
AW760858 weakly similar to GP|15810014|gb| At2g44770/F16B22.26 {... 32 0.22
AW233793 weakly similar to GP|27450574|gb pollen-specific calmod... 27 7.2
TC214866 homologue to UP|XTHB_PHAAN (Q8LNZ5) Probable xyloglucan... 27 7.2
TC225009 homologue to UP|Q9FYF3 (Q9FYF3) F1N21.22, partial (19%) 27 9.4
TC215543 similar to UP|Q8LDN3 (Q8LDN3) F-box protein AtFBL5, par... 27 9.4
TC222869 similar to UP|Q8W529 (Q8W529) Methionine synthase (Frag... 27 9.4
AI737026 similar to GP|12836895|gb| senescence-associated protei... 27 9.4
>TC217971 similar to UP|Q940L9 (Q940L9) At2g44770/F16B22.26, partial (89%)
Length = 1446
Score = 229 bits (583), Expect(2) = e-107
Identities = 106/122 (86%), Positives = 115/122 (93%)
Frame = +1
Query: 24 YHSSSAEFVTGSTAWIGRGLSCVCAQRRESDARLSFDLTPYQEECLQRLQSRIDVPYDSS 83
++ +AEFVTGSTAW+GRGLSCVCAQRRESDAR SFDLTP QEECLQRLQSRID+PYD S
Sbjct: 187 FYFVTAEFVTGSTAWLGRGLSCVCAQRRESDARPSFDLTPAQEECLQRLQSRIDIPYDGS 366
Query: 84 IPEHQASLRALWNAAFPEEELNGLISEQWKDMGWQGKDPSTDFRGGGYISLENLLFFARN 143
+PEHQ +LR LWNAAFPEEEL+GLISEQWKDMGWQGKDPSTDFRGGG+ISLENLLFFARN
Sbjct: 367 VPEHQDALRDLWNAAFPEEELHGLISEQWKDMGWQGKDPSTDFRGGGFISLENLLFFARN 546
Query: 144 FP 145
FP
Sbjct: 547 FP 552
Score = 176 bits (447), Expect(2) = e-107
Identities = 94/126 (74%), Positives = 100/126 (78%), Gaps = 1/126 (0%)
Frame = +2
Query: 137 LLFFARN-FPIFCGSRKEIVQCGSTRLRLLVLTLHSCLFKC*ILKQLSHGHWWEQLS*NS 195
LL +RN F IF GSRKEI QCG+ L LLVLT HSCLF+C ILKQ + GHWWEQLS*N
Sbjct: 536 LLGISRNPFKIFYGSRKEIGQCGNIHLLLLVLTSHSCLFRCWILKQSNLGHWWEQLS*NF 715
Query: 196 LQKMSRPLIFSIA*HSS*WTINGFPCTRHTWILIQ**NLHAVSWRKNFSLKTLHN*KMYP 255
LQKM + LIFSIA*HS *W NGF C HTWILIQ**NLHAVSWRK+F LKT *KMYP
Sbjct: 716 LQKMIQLLIFSIA*HSR*WINNGFLCVLHTWILIQ**NLHAVSWRKSFYLKT*CG*KMYP 895
Query: 256 HTNFSH 261
HTNFSH
Sbjct: 896 HTNFSH 913
>TC229862 similar to UP|Q940L9 (Q940L9) At2g44770/F16B22.26, partial (89%)
Length = 1295
Score = 205 bits (521), Expect = 2e-53
Identities = 96/118 (81%), Positives = 105/118 (88%)
Frame = +3
Query: 28 SAEFVTGSTAWIGRGLSCVCAQRRESDARLSFDLTPYQEECLQRLQSRIDVPYDSSIPEH 87
+AE V GS AW+GRGLSCVC QRR+SDA +FDLT QEECL R+Q RIDVPYDSSI EH
Sbjct: 66 TAEVVVGSAAWLGRGLSCVCVQRRDSDASNTFDLTLAQEECLHRIQRRIDVPYDSSIIEH 245
Query: 88 QASLRALWNAAFPEEELNGLISEQWKDMGWQGKDPSTDFRGGGYISLENLLFFARNFP 145
Q +LRALWNAAFPEEEL+GLISEQWK+MGWQGKDPSTDFRGGG+ISLEN LFFARNFP
Sbjct: 246 QDALRALWNAAFPEEELHGLISEQWKEMGWQGKDPSTDFRGGGFISLENFLFFARNFP 419
Score = 139 bits (349), Expect = 2e-33
Identities = 79/125 (63%), Positives = 90/125 (71%), Gaps = 1/125 (0%)
Frame = +1
Query: 136 NLLFFARN-FPIFCGSRKEIVQCGSTRLRLLVLTLHSCLFKC*ILKQLSHGHWWEQLS*N 194
+LL +RN F SRKEI QCG+T L+LLVLT H+C +KC ILKQ SH W EQL *N
Sbjct: 400 SLLGISRNPSRFFYASRKEIDQCGNTHLQLLVLTSHTC*YKCLILKQSSHAIWLEQLF*N 579
Query: 195 SLQKMSRPLIFSIA*HSS*WTINGFPCTRHTWILIQ**NLHAVSWRKNFSLKTLHN*KMY 254
LQKM + LIFSIA* SS*W ING C HTWIL+Q** H VS RK+FSLKT K+Y
Sbjct: 580 FLQKMGQLLIFSIA*LSS*WIINGLLCVPHTWILMQ**KKHDVS*RKSFSLKT*PGLKIY 759
Query: 255 PHTNF 259
PHTN+
Sbjct: 760 PHTNY 774
>TC217970
Length = 711
Score = 155 bits (393), Expect = 1e-38
Identities = 75/84 (89%), Positives = 78/84 (92%)
Frame = +3
Query: 1 MDDRGGSFVAVRRISQGLDRGNAYHSSSAEFVTGSTAWIGRGLSCVCAQRRESDARLSFD 60
MDDRGGSFVAVRRISQGLDR N HSSSAEFVTGSTAW+GRGLSCVCAQRRESDAR SFD
Sbjct: 459 MDDRGGSFVAVRRISQGLDRSNTCHSSSAEFVTGSTAWLGRGLSCVCAQRRESDARPSFD 638
Query: 61 LTPYQEECLQRLQSRIDVPYDSSI 84
LTP QEECLQRLQSRID+PYD S+
Sbjct: 639 LTPAQEECLQRLQSRIDIPYDGSV 710
>TC216136
Length = 1291
Score = 121 bits (304), Expect(2) = 4e-33
Identities = 61/118 (51%), Positives = 80/118 (67%), Gaps = 1/118 (0%)
Frame = +3
Query: 29 AEFVTGSTAWIGRGLSCVCAQRRESDARL-SFDLTPYQEECLQRLQSRIDVPYDSSIPEH 87
A V GS + +GR LS A + R+ L+P QEE L+ L+ R++VP+D S EH
Sbjct: 417 ANLVLGSGSILGRLLSFPSAALNMQNNRMFPPSLSPLQEERLRNLRQRLEVPFDGSKAEH 596
Query: 88 QASLRALWNAAFPEEELNGLISEQWKDMGWQGKDPSTDFRGGGYISLENLLFFARNFP 145
Q +L+ LW A+P+ EL L S+ WK+MGWQG DPSTDFRGGG+ISLENL+FFA +P
Sbjct: 597 QDALKQLWKLAYPDRELPSLKSDLWKEMGWQGSDPSTDFRGGGFISLENLIFFAMKYP 770
Score = 37.4 bits (85), Expect(2) = 4e-33
Identities = 34/108 (31%), Positives = 48/108 (43%)
Frame = +1
Query: 147 FCGSRKEIVQCGSTRLRLLVLTLHSCLFKC*ILKQLSHGHWWEQLS*NSLQKMSRPLIFS 206
+C ++ E G+T L+ L C +C I KQ H E + N L+KM LI
Sbjct: 787 YCINKMEPELSGNTHLQ*LESIFLLC*HRCWIFKQGFHLLCPEFVFSNCLKKMKWHLISF 966
Query: 207 IA*HSS*WTINGFPCTRHTWILIQ**NLHAVSWRKNFSLKTLHN*KMY 254
*W ++G RH W L+ *+ S + LKT *K+Y
Sbjct: 967 FVLPFK*WMLSG*QSVRHIWNLMMF*DPQERS*SVSLVLKTSSA*KIY 1110
>BG789642
Length = 409
Score = 122 bits (307), Expect = 1e-28
Identities = 62/91 (68%), Positives = 69/91 (75%)
Frame = +3
Query: 1 MDDRGGSFVAVRRISQGLDRGNAYHSSSAEFVTGSTAWIGRGLSCVCAQRRESDARLSFD 60
MDDRG SF+ VRRI G HS+SAE V GS AW+GRGLSCVC QRR+SDA +FD
Sbjct: 147 MDDRGSSFITVRRIPHG----ETCHSNSAEVVVGSAAWLGRGLSCVCVQRRDSDASNTFD 314
Query: 61 LTPYQEECLQRLQSRIDVPYDSSIPEHQASL 91
LT QEECLQR+Q RIDVPYDSSI EHQ +L
Sbjct: 315 LTLAQEECLQRIQRRIDVPYDSSIIEHQDAL 407
>CD404946
Length = 353
Score = 82.8 bits (203), Expect(2) = 1e-19
Identities = 36/48 (75%), Positives = 41/48 (85%)
Frame = -3
Query: 98 AFPEEELNGLISEQWKDMGWQGKDPSTDFRGGGYISLENLLFFARNFP 145
+FP L GLIS+QWKDMGWQG +PSTDFRG G+ISLENLLFFAR +P
Sbjct: 351 SFPNVSLEGLISDQWKDMGWQGPNPSTDFRGCGFISLENLLFFARKYP 208
Score = 30.8 bits (68), Expect(2) = 1e-19
Identities = 20/62 (32%), Positives = 30/62 (48%)
Frame = -1
Query: 144 FPIFCGSRKEIVQCGSTRLRLLVLTLHSCLFKC*ILKQLSHGHWWEQLS*NSLQKMSRPL 203
F C + E Q G+ L LL L H C ++C I Q SH + + + +KM + L
Sbjct: 200 FTSCC*RKMETEQPGNIHLLLLALIYHLC*YRCWIYAQKSHVVFQA*ILSSY*EKMKKHL 21
Query: 204 IF 205
+F
Sbjct: 20 MF 15
>BE802287 weakly similar to GP|9828622|gb|A F1N21.22 {Arabidopsis thaliana},
partial (23%)
Length = 387
Score = 86.7 bits (213), Expect = 1e-17
Identities = 43/86 (50%), Positives = 61/86 (70%), Gaps = 2/86 (2%)
Frame = +3
Query: 25 HSSSAEFVTGS--TAWIGRGLSCVCAQRRESDARLSFDLTPYQEECLQRLQSRIDVPYDS 82
H+S GS +AWIG+GL+CVC +R+ + R+ LTP QEE L+RL+ R+ V +D+
Sbjct: 129 HASDHATTCGSPASAWIGKGLTCVCFKRKGNCQRICISLTPLQEERLRRLKRRMKVYFDA 308
Query: 83 SIPEHQASLRALWNAAFPEEELNGLI 108
S EHQ +LRALW+A+FP++EL LI
Sbjct: 309 SKLEHQEALRALWSASFPDQELQSLI 386
>TC208530
Length = 579
Score = 80.1 bits (196), Expect = 9e-16
Identities = 36/51 (70%), Positives = 41/51 (79%)
Frame = +1
Query: 30 EFVTGSTAWIGRGLSCVCAQRRESDARLSFDLTPYQEECLQRLQSRIDVPY 80
+ V GS AW+GRGLSCVC QRR+SD +FDLT QEECLQR+Q RIDVPY
Sbjct: 427 QVVAGSAAWLGRGLSCVCVQRRDSDVSNTFDLTLAQEECLQRIQRRIDVPY 579
Score = 38.9 bits (89), Expect = 0.002
Identities = 23/51 (45%), Positives = 28/51 (54%)
Frame = +3
Query: 1 MDDRGGSFVAVRRISQGLDRGNAYHSSSAEFVTGSTAWIGRGLSCVCAQRR 51
MDDRG SFVAVRRI Q G H +SA S R + C+C ++R
Sbjct: 354 MDDRGSSFVAVRRIPQ----GETCHPNSAGCGRVSCMARSRSVLCLCTKKR 494
>TC216135
Length = 685
Score = 75.1 bits (183), Expect = 3e-14
Identities = 39/91 (42%), Positives = 57/91 (61%), Gaps = 1/91 (1%)
Frame = +3
Query: 29 AEFVTGSTAWIGRGLSCVCAQ-RRESDARLSFDLTPYQEECLQRLQSRIDVPYDSSIPEH 87
A V GS + +GR LS A +++ L L+P QE+ L+ L+ R++VP+D S EH
Sbjct: 411 ANVVLGSGSILGRLLSFPSAALNMQNNRMLPPSLSPLQEDRLRNLRQRLEVPFDGSKAEH 590
Query: 88 QASLRALWNAAFPEEELNGLISEQWKDMGWQ 118
Q +L+ LW A+P+ EL L S+ WK+MGWQ
Sbjct: 591 QDALKLLWKLAYPDRELPSLKSDLWKEMGWQ 683
>BU764054 weakly similar to GP|9828622|gb|A F1N21.22 {Arabidopsis thaliana},
partial (17%)
Length = 472
Score = 75.1 bits (183), Expect = 3e-14
Identities = 35/65 (53%), Positives = 49/65 (74%)
Frame = +3
Query: 34 GSTAWIGRGLSCVCAQRRESDARLSFDLTPYQEECLQRLQSRIDVPYDSSIPEHQASLRA 93
GS AWIG+GL+CVC +R+ + R+ LTP QEE L+RL+ R+ V +D+S EHQ +LRA
Sbjct: 276 GSPAWIGKGLTCVCFKRKGNCQRICISLTPLQEERLRRLKRRMKVYFDASKLEHQEALRA 455
Query: 94 LWNAA 98
LW+A+
Sbjct: 456 LWSAS 470
>BE806603
Length = 413
Score = 67.8 bits (164), Expect = 5e-12
Identities = 31/36 (86%), Positives = 33/36 (91%)
Frame = +2
Query: 66 EECLQRLQSRIDVPYDSSIPEHQASLRALWNAAFPE 101
EECLQR+Q RIDVPYDSSI EHQ +LRALWNAAFPE
Sbjct: 305 EECLQRIQRRIDVPYDSSIIEHQDALRALWNAAFPE 412
Score = 33.1 bits (74), Expect = 0.13
Identities = 15/17 (88%), Positives = 15/17 (88%)
Frame = +1
Query: 1 MDDRGGSFVAVRRISQG 17
MDDRG SFVAVRRI QG
Sbjct: 232 MDDRGSSFVAVRRIPQG 282
>AW704329 weakly similar to GP|9828622|gb|A F1N21.22 {Arabidopsis thaliana},
partial (10%)
Length = 359
Score = 35.0 bits (79), Expect(2) = 1e-05
Identities = 17/33 (51%), Positives = 23/33 (69%)
Frame = +3
Query: 56 RLSFDLTPYQEECLQRLQSRIDVPYDSSIPEHQ 88
R+ LTP QEE L+RL+ R+ V +D+S EHQ
Sbjct: 255 RICISLTPLQEERLRRLKRRMKVYFDASKLEHQ 353
Score = 30.8 bits (68), Expect(2) = 1e-05
Identities = 10/17 (58%), Positives = 16/17 (93%)
Frame = +1
Query: 35 STAWIGRGLSCVCAQRR 51
++AWIG+GL+CVC +R+
Sbjct: 193 ASAWIGKGLTCVCFKRK 243
>TC216137
Length = 519
Score = 35.4 bits (80), Expect = 0.026
Identities = 28/80 (35%), Positives = 38/80 (47%)
Frame = +2
Query: 175 KC*ILKQLSHGHWWEQLS*NSLQKMSRPLIFSIA*HSS*WTINGFPCTRHTWILIQ**NL 234
+C I KQ H E + N L++M LI * *W ++G P RH W L+ *+
Sbjct: 5 RCWIFKQGFHLLRLEFIFSNCLKRMKWHLISFFV*PFK*WMLSG*PSVRHIWNLMMF*SP 184
Query: 235 HAVSWRKNFSLKTLHN*KMY 254
S + LKT *K+Y
Sbjct: 185 QERS*SVSLLLKTFPA*KIY 244
>AW760858 weakly similar to GP|15810014|gb| At2g44770/F16B22.26 {Arabidopsis
thaliana}, partial (23%)
Length = 421
Score = 32.3 bits (72), Expect = 0.22
Identities = 16/46 (34%), Positives = 23/46 (49%)
Frame = +2
Query: 197 QKMSRPLIFSIA*HSS*WTINGFPCTRHTWILIQ**NLHAVSWRKN 242
+KM + +F *H *W NG+ C TWIL +WR++
Sbjct: 284 EKMKKHSMFYTV*HLR*WMHNGWLCMLPTWILTMFYKQQECNWRES 421
>AW233793 weakly similar to GP|27450574|gb pollen-specific calmodulin-binding
protein {Arabidopsis thaliana}, partial (16%)
Length = 393
Score = 27.3 bits (59), Expect = 7.2
Identities = 14/32 (43%), Positives = 18/32 (55%)
Frame = +3
Query: 160 TRLRLLVLTLHSCLFKC*ILKQLSHGHWWEQL 191
++LRLL LTL F +L HWW+QL
Sbjct: 159 SKLRLLPLTLRGLTFSPKLLS-----HWWQQL 239
>TC214866 homologue to UP|XTHB_PHAAN (Q8LNZ5) Probable xyloglucan
endotransglucosylase/hydrolase protein B precursor
(VaXTH2) , complete
Length = 1124
Score = 27.3 bits (59), Expect = 7.2
Identities = 13/40 (32%), Positives = 19/40 (47%)
Frame = -3
Query: 45 CVCAQRRESDARLSFDLTPYQEECLQRLQSRIDVPYDSSI 84
C C + E D+ LS L + CLQ ++ VP + I
Sbjct: 462 CFCGIKPEKDSLLSVTLASGEHICLQNVRLSCSVPQELKI 343
>TC225009 homologue to UP|Q9FYF3 (Q9FYF3) F1N21.22, partial (19%)
Length = 414
Score = 26.9 bits (58), Expect = 9.4
Identities = 11/14 (78%), Positives = 13/14 (92%)
Frame = -1
Query: 131 YISLENLLFFARNF 144
+ISLENLLFFA+ F
Sbjct: 183 FISLENLLFFAKTF 142
>TC215543 similar to UP|Q8LDN3 (Q8LDN3) F-box protein AtFBL5, partial (46%)
Length = 549
Score = 26.9 bits (58), Expect = 9.4
Identities = 16/56 (28%), Positives = 25/56 (44%), Gaps = 3/56 (5%)
Frame = +2
Query: 61 LTPY---QEECLQRLQSRIDVPYDSSIPEHQASLRALWNAAFPEEELNGLISEQWK 113
L PY Q CL + S +VP+ +P+H L+ E++ G+ WK
Sbjct: 275 LCPYNR*QCNCLGKQMSSSEVPWAVLLPKHNRQGNVLFGTKQIEQQDVGVCERWWK 442
>TC222869 similar to UP|Q8W529 (Q8W529) Methionine synthase (Fragment),
partial (19%)
Length = 587
Score = 26.9 bits (58), Expect = 9.4
Identities = 14/48 (29%), Positives = 24/48 (49%), Gaps = 3/48 (6%)
Frame = -2
Query: 27 SSAEFVTGSTAWIGRGLSCVCAQRRESD---ARLSFDLTPYQEECLQR 71
S +F GS AWI + C C++R ++ R+ + + EC +R
Sbjct: 205 SKLKFSLGSHAWITNNMRCHCSRRSGTNREAKRIG*ERESAERECKRR 62
>AI737026 similar to GP|12836895|gb| senescence-associated protein {Ipomoea
batatas}, partial (19%)
Length = 412
Score = 26.9 bits (58), Expect = 9.4
Identities = 13/37 (35%), Positives = 22/37 (59%), Gaps = 1/37 (2%)
Frame = -3
Query: 61 LTPYQEECLQ-RLQSRIDVPYDSSIPEHQASLRALWN 96
+ P Q++C + L ++ +PY S +PE Q S LW+
Sbjct: 230 IKPLQDQCNRFHLPQKVLMPYSSMLPEEQQSF--LWS 126
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.337 0.145 0.501
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,589,195
Number of Sequences: 63676
Number of extensions: 236480
Number of successful extensions: 1552
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 1544
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1550
length of query: 261
length of database: 12,639,632
effective HSP length: 95
effective length of query: 166
effective length of database: 6,590,412
effective search space: 1094008392
effective search space used: 1094008392
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC149129.2


