

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149129.14 - phase: 0 /pseudo
(624 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
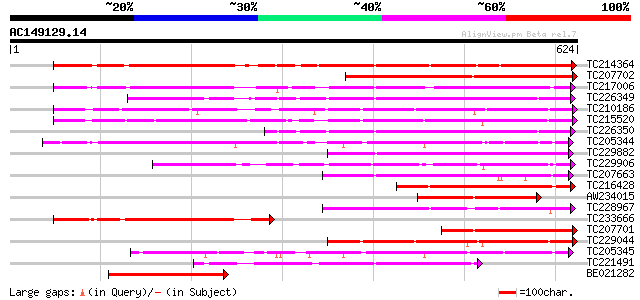
Score E
Sequences producing significant alignments: (bits) Value
TC214364 UP|Q6WNU4 (Q6WNU4) Subtilisin-like protease, complete 463 e-131
TC207702 weakly similar to UP|Q8S896 (Q8S896) Subtilisin-like se... 385 e-107
TC217006 UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C1, complete 346 2e-95
TC226349 similar to PIR|JC7519|JC7519 subtilisin-like serine pro... 338 3e-93
TC210186 UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease, complete 330 9e-91
TC215520 GB|AAG38994.1|11611651|AF160513 subtilisin-type proteas... 303 1e-82
TC226350 similar to PIR|JC7519|JC7519 subtilisin-like serine pro... 257 9e-69
TC205344 similar to UP|Q9SZV5 (Q9SZV5) Proteinase-like protein (... 254 8e-68
TC229882 weakly similar to UP|Q75I27 (Q75I27) Putaive subtilisin... 230 1e-60
TC229906 weakly similar to UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like pr... 225 4e-59
TC207663 similar to UP|P93204 (P93204) SBT1 protein (Subtilisin-... 225 5e-59
TC216428 similar to UP|Q9AX30 (Q9AX30) Subtilisin-like protease,... 224 7e-59
AW234015 223 3e-58
TC228967 similar to GB|BAB09208.1|9758669|AB012245 subtilisin-li... 215 5e-56
TC233666 weakly similar to UP|Q9FJF3 (Q9FJF3) Serine protease-li... 203 2e-52
TC207701 similar to UP|Q84TR6 (Q84TR6) Subtilase, partial (6%) 202 3e-52
TC229044 similar to UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease,... 202 3e-52
TC205345 similar to UP|Q9SZV5 (Q9SZV5) Proteinase-like protein (... 201 1e-51
TC221491 similar to UP|P93204 (P93204) SBT1 protein (Subtilisin-... 196 2e-50
BE021282 similar to GP|21593457|gb| subtilisin-like serine prote... 185 5e-47
>TC214364 UP|Q6WNU4 (Q6WNU4) Subtilisin-like protease, complete
Length = 2667
Score = 463 bits (1192), Expect = e-131
Identities = 266/579 (45%), Positives = 353/579 (60%), Gaps = 4/579 (0%)
Frame = +1
Query: 49 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 108
TAGGN V S+FG G+GT KGGSP +RV YKVCW D C+ AD+L+A D A
Sbjct: 721 TAGGNMVARVSVFGQGHGTAKGGSPMARVAAYKVCWPPVAGDE----CFDADILAAFDLA 888
Query: 109 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 168
I DGVD++SVS+GG SS+ F D ++IG+F A + +++V SAGN GP + N+A
Sbjct: 889 IHDGVDVLSVSLGGSSST----FFKDSVAIGSFHAAKRGVVVVCSAGNSGPAEATAENLA 1056
Query: 169 PWVFTVAASTIDRDFSSTITIGNK-TVTGASLFVNLPPNQSFTLVDSIDAKFANVTNQDA 227
PW TVAAST+DR F + + +GN T G SL ++ + ++ + DAK A+ +DA
Sbjct: 1057 PWHVTVAASTMDRQFPTYVVLGNDITFKGESLSATKLAHKFYPIIKATDAKLASARAEDA 1236
Query: 228 RFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKGM 287
C+ GTLDP+K GKIV C+ ++ R V +G +A AGA GM
Sbjct: 1237 VLCQNGTLDPNKAKGKIVVCL----------RGINAR--------VDKGEQAFLAGAVGM 1362
Query: 288 ILRNQPKFNGKTLLAESNVL--STINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQPKT 345
+L N K G ++A+ +VL S IN+ D G ++ T V I + PKT
Sbjct: 1363 VLAND-KTTGNEIIADPHVLPASHINFTD------GSAVFKYINSTKFPVAYI--THPKT 1515
Query: 346 SYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPF 405
KPAP MA+FSS+GPN + P ILKPD+TAPGV+++AAY+ +N V D RR PF
Sbjct: 1516 QLDTKPAPFMAAFSSKGPNTIVPEILKPDITAPGVSVIAAYTEAQGPTNQVFDKRR-IPF 1692
Query: 406 NIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLAN 465
N GTSMSCPHV+G GL++ L+P WS AAIKSAIMTTAT DN + + +A D A
Sbjct: 1693 NSVSGTSMSCPHVSGIVGLLRALYPTWSTAAIKSAIMTTATTLDNEVEPLLNATDGK-AT 1869
Query: 466 PFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIHSI 525
PF+YG+GH+QPN AMDPGLVYD+++ DYLNFLCA GY++ IS + C S+
Sbjct: 1870 PFSYGAGHVQPNRAMDPGLVYDITIDDYLNFLCALGYNETQISVFTEG--PYKCRKKFSL 2043
Query: 526 NDLNYPSITLPNLGLNAVNVTRIVTNVGPPSTYFAKVQLP-GYNIVVVPDSLTFKKNGEK 584
+LNYPSIT+P L +V VTR + NVG P TY A VQ P G + V P L FK GE+
Sbjct: 2044 LNLNYPSITVPKLS-GSVTVTRTLKNVGSPGTYIAHVQNPYGITVSVKPSILKFKNVGEE 2220
Query: 585 KKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTVQ 623
K F++ +A Y FG+L W++GKH V SP+ V+
Sbjct: 2221 KSFKLTFKAMQGKATNNYAFGKLIWSDGKHYVTSPIVVK 2337
>TC207702 weakly similar to UP|Q8S896 (Q8S896) Subtilisin-like serine
protease AIR3 (Fragment), partial (20%)
Length = 1025
Score = 385 bits (989), Expect = e-107
Identities = 193/256 (75%), Positives = 215/256 (83%), Gaps = 1/256 (0%)
Frame = +2
Query: 370 ILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQGTSMSCPHVAGTAGLIKTLH 429
ILKPDVTAPGVNILAAYS AS SNL+ DNRRGF FN+ QGTS+SCPHVAG AGLIKTLH
Sbjct: 5 ILKPDVTAPGVNILAAYSELASASNLLVDNRRGFKFNVLQGTSVSCPHVAGIAGLIKTLH 184
Query: 430 PNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPFAYGSGHIQPNTAMDPGLVYDLS 489
PNWSPAAIKSAIMTTAT DNTN+ I+DA D +A+ FAYGSGH+QP+ A+DPGLVYDLS
Sbjct: 185 PNWSPAAIKSAIMTTATTLDNTNRPIQDAFDNKVADAFAYGSGHVQPDLAIDPGLVYDLS 364
Query: 490 VVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIHSINDLNYPSITLPNLGLNAVNVTRIV 549
+ DYLNFLCA+GY Q+LIS LN N TF C G HS+ DLNYPSITLPNLGL V +TR V
Sbjct: 365 LADYLNFLCASGYDQQLISA-LNFNGTFICKGSHSVTDLNYPSITLPNLGLKPVTITRTV 541
Query: 550 TNVGPPSTYFAKVQLP-GYNIVVVPDSLTFKKNGEKKKFQVIVQARSVTPRGRYQFGELQ 608
TNVGPP+TY A V P GY IVVVP SLTF K GEKKKFQVIVQA SVT R +YQFG+L+
Sbjct: 542 TNVGPPATYTANVHSPAGYTIVVVPRSLTFTKIGEKKKFQVIVQASSVTTRRKYQFGDLR 721
Query: 609 WTNGKHIVRSPVTVQR 624
WT+GKHIVRSP+TV+R
Sbjct: 722 WTDGKHIVRSPITVKR 769
>TC217006 UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C1, complete
Length = 2422
Score = 346 bits (888), Expect = 2e-95
Identities = 236/591 (39%), Positives = 306/591 (50%), Gaps = 17/591 (2%)
Frame = +3
Query: 49 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 108
TA GN V AS+ G+G GT +GG K+R+ YKVCW DG C AD+L+A D A
Sbjct: 648 TAAGNPVSTASMLGLGQGTSRGGVTKARIAVYKVCWF----DG----CTDADILAAFDDA 803
Query: 109 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 168
I+DGVDII+VS+GG S N+ F D I+IGAF A +L V SAGN GP P S++N +
Sbjct: 804 IADGVDIITVSLGGFSDENY---FRDGIAIGAFHAVRNGVLTVTSAGNSGPRPSSLSNFS 974
Query: 169 PWVFTVAASTIDRDFSSTITIGNK-TVTGASLFVNLPPNQSFTLVDSIDA--KFANVTNQ 225
PW +VAASTIDR F + + +GNK T G S+ + + ++ DA K +
Sbjct: 975 PWSISVAASTIDRKFVTKVELGNKITYEGTSINTFDLKGELYPIIYGGDAPNKGEGIDGS 1154
Query: 226 DARFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAK 285
+R+C G+LD V GKIV C S S+ AGA
Sbjct: 1155 SSRYCSSGSLDKKLVKGKIVLC-----------------------ESRSKALGPFDAGAV 1265
Query: 286 GMILRNQP--------KFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIK 337
G +++ Q G L + + YD TR I TD K I
Sbjct: 1266 GALIQGQGFRDLPPSLPLPGSYLALQDGA----SVYDYINSTRTPIATIFKTDETKDTI- 1430
Query: 338 IRMSQPKTSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVT 397
APV+ASFSSRGPN V P ILKPD+ APGV+ILA++S + S++
Sbjct: 1431 --------------APVVASFSSRGPNIVTPEILKPDLVAPGVSILASWSPASPPSDVEG 1568
Query: 398 DNRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRD 457
DNR FNI GTSM+CPHV+G A +K+ HP WSPAAI+SA+MTTA L+ +
Sbjct: 1569 DNRT-LNFNIISGTSMACPHVSGAAAYVKSFHPTWSPAAIRSALMTTAKQLSPKTHLLAE 1745
Query: 458 AIDKTLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTF 517
FAYG+G I P+ A+ PGLVYD +DY+ FLC GYS R + + N +
Sbjct: 1746 ---------FAYGAGQIDPSKAVYPGLVYDAGEIDYVRFLCGQGYSTRTLQLITGDNSSC 1898
Query: 518 TCSGIHSINDLNYPSITL--PNLGLNAV--NVTRIVTNVG-PPSTYFAKVQLP-GYNIVV 571
+ S DLNY S L P N+V + R VTNVG P STY A V P G I V
Sbjct: 1899 PETKNGSARDLNYASFALFVPPYNSNSVSGSFNRTVTNVGSPKSTYKATVTSPKGLKIEV 2078
Query: 572 VPDSLTFKKNGEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTV 622
P L F +K+ F + + + G G L W +GK+ VRSP+ V
Sbjct: 2079 NPSVLPFTSLNQKQTFVLTITGKL---EGPIVSGSLVWDDGKYQVRSPIVV 2222
>TC226349 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(56%)
Length = 1773
Score = 338 bits (868), Expect = 3e-93
Identities = 214/500 (42%), Positives = 281/500 (55%), Gaps = 7/500 (1%)
Frame = +1
Query: 130 EIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVAPWVFTVAASTIDRDFSSTITI 189
+ + D ++IGAF A K IL+ SAGN GP P S++NVAPW+ TV A T+DRDF + + +
Sbjct: 1 DYYRDSVAIGAFSAMEKGILVSCSAGNSGPGPYSLSNVAPWITTVGAGTLDRDFPAYVAL 180
Query: 190 GNK-TVTGASLFV-NLPPNQSFTLVDSIDAKFANVTN--QDARFCKPGTLDPSKVSGKIV 245
GN +G SL+ N P+ S LV + NV+N + C GTL P KV+GKIV
Sbjct: 181 GNGLNFSGVSLYRGNALPDSSLPLVYA-----GNVSNGAMNGNLCITGTLSPEKVAGKIV 345
Query: 246 ECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKGMILRNQPKFNGKTLLAESN 305
C R L T V +G SAGA GM+L N NG+ L+A+++
Sbjct: 346 LC---------------DRGL---TARVQKGSVVKSAGALGMVLSNTAA-NGEELVADAH 468
Query: 306 VLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRMSQPKTSYRRKPAPVMASFSSRGPNQ 365
+L K G +I K +KI K + P+PV+A+FSSRGPN
Sbjct: 469 LLPATAVGQK----AGDAIKKYLVSDAKPTVKIFFEGTKVGIQ--PSPVVAAFSSRGPNS 630
Query: 366 VQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQGTSMSCPHVAGTAGLI 425
+ P ILKPD+ APGVNILA +S + L DNRR FNI GTSMSCPHV+G A LI
Sbjct: 631 ITPQILKPDLIAPGVNILAGWSKAVGPTGLPVDNRR-VDFNIISGTSMSCPHVSGLAALI 807
Query: 426 KTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPFAYGSGHIQPNTAMDPGLV 485
K+ HP+WSPAA++SA+MTTA T + ++D+ + PF +GSGH+ P A++PGLV
Sbjct: 808 KSAHPDWSPAAVRSALMTTAYTVYKTGEKLQDSATGKPSTPFDHGSGHVDPVAALNPGLV 987
Query: 486 YDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIHSINDLNYPSI-TLPNLGLNAVN 544
YDL+V DYL FLCA YS ISTL +S+ DLNYPS L + V
Sbjct: 988 YDLTVDDYLGFLCALNYSAAEISTLAKRKFQCDAGKQYSVTDLNYPSFAVLFESSGSVVK 1167
Query: 545 VTRIVTNVGPPSTYFAKV--QLPGYNIVVVPDSLTFKKNGEKKKFQVIVQARSVTPRGRY 602
TR +TNVGP TY A V I V P L+FK+N EKK F V +
Sbjct: 1168HTRTLTNVGPAGTYKASVTSDTASVKISVEPQVLSFKEN-EKKTFTVTFSSSGSPQHTEN 1344
Query: 603 QFGELQWTNGKHIVRSPVTV 622
FG ++W++GKH+V SP++V
Sbjct: 1345AFGRVEWSDGKHLVGSPISV 1404
>TC210186 UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease, complete
Length = 2401
Score = 330 bits (847), Expect = 9e-91
Identities = 219/593 (36%), Positives = 324/593 (53%), Gaps = 17/593 (2%)
Frame = +1
Query: 49 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 108
TA G V AS +G+ G KGGSP+SR+ Y+VC ++ C G+ +L+A D A
Sbjct: 718 TAAGVMVTNASYYGVATGCAKGGSPESRLAVYRVC--------SNFGCRGSSILAAFDDA 873
Query: 109 ISDGVDIISVSVGGRSSSNFE-EIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNV 167
I+DGVD++SVS+G +S+ F ++ +D IS+GAF A IL+V SAGN GP+ ++ N
Sbjct: 874 IADGVDLLSVSLG--ASTGFRPDLTSDPISLGAFHAMEHGILVVCSAGNDGPSSYTLVND 1047
Query: 168 APWVFTVAASTIDRDFSSTITIG-NKTVTGASLFVNLPP---NQSFTLVDSIDAKFANVT 223
APW+ TVAASTIDR+F S I +G NK + G + +NL P + + L+ AK + +
Sbjct: 1048 APWILTVAASTIDRNFLSNIVLGDNKIIKGKA--INLSPLSNSPKYPLIYGESAKANSTS 1221
Query: 224 NQDARFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAG 283
+AR C P +LD +KV GKIV C + + S ++ +
Sbjct: 1222 LVEARQCHPNSLDGNKVKGKIVVCDDK-------------------NDKYSTRKKVATVK 1344
Query: 284 AKGMILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKS----VIKIR 339
A G I G + + N NY D G++ I S V I
Sbjct: 1345 AVGGI--------GLVHITDQNEAIASNYGDFPATVISSKDGVTILQYINSTSNPVATIL 1500
Query: 340 MSQPKTSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDN 399
+ Y KPAP++ +FSSRGP+ + ILKPD+ APGVNILAA+ + + +V
Sbjct: 1501 ATTSVLDY--KPAPLVPNFSSRGPSSLSSNILKPDIAAPGVNILAAW--IGNGTEVVPKG 1668
Query: 400 RRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAI 459
++ + I GTSM+CPHV+G A +KT +P WS ++IKSAIMT+A I+ N K
Sbjct: 1669 KKPSLYKIISGTSMACPHVSGLASSVKTRNPTWSASSIKSAIMTSA-IQSNNLKAPITTE 1845
Query: 460 DKTLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTL-----LNPN 514
++A P+ YG+G + + + PGLVY+ S VDYLNFLC G++ + + N N
Sbjct: 1846 SGSVATPYDYGAGEMTTSEPLQPGLVYETSSVDYLNFLCYIGFNVTTVKVISKTVPRNFN 2025
Query: 515 MTFTCSGIHSINDLNYPSITLPNLGLNAVNVTRIVTNVG--PPSTYFAKVQLP-GYNIVV 571
S H I+++NYPSI + G AVN++R VTNVG + Y V P G ++ +
Sbjct: 2026 CPKDLSSDH-ISNINYPSIAINFSGKRAVNLSRTVTNVGEDDETVYSPIVDAPSGVHVTL 2202
Query: 572 VPDSLTFKKNGEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTVQR 624
P+ L F K+ +K ++VI + ++T FG + W+NGK++VRSP + +
Sbjct: 2203 TPNKLRFTKSSKKLSYRVIFSS-TLTSLKEDLFGSITWSNGKYMVRSPFVLTK 2358
>TC215520 GB|AAG38994.1|11611651|AF160513 subtilisin-type protease precursor
{Glycine max;} , complete
Length = 2392
Score = 303 bits (777), Expect = 1e-82
Identities = 216/587 (36%), Positives = 313/587 (52%), Gaps = 11/587 (1%)
Frame = +2
Query: 49 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 108
TA G V GAS +G+ GT +GGSP+SR+ YKVC G C G+ +L+ D A
Sbjct: 692 TAVGVPVSGASFYGLAAGTARGGSPESRLAVYKVC-------GAFGSCPGSAILAGFDDA 850
Query: 109 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 168
I DGVDI+S+S+GG + ++ TD I+IGAF + + IL+V +AGN G P +V N A
Sbjct: 851 IHDGVDILSLSLGGFGGTK-TDLTTDPIAIGAFHSVQRGILVVCAAGNDGE-PFTVLNDA 1024
Query: 169 PWVFTVAASTIDRDFSSTITIG-NKTVTGASL-FVNLPPNQSFTLVDSIDAKFANVTN-Q 225
PW+ TVAASTIDRD S + +G N+ V G ++ F L + + ++ + A AN++N
Sbjct: 1025 PWILTVAASTIDRDLQSDVVLGNNQVVKGRAINFSPLLNSPDYPMIYAESAARANISNIT 1204
Query: 226 DARFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAK 285
DAR C P +LDP KV GKIV C G+ +T E + ++ A + +G+
Sbjct: 1205 DARQCHPDSLDPKKVIGKIVVCDGKNDIYYSTDEKI---VIVKALGGIGLVHITDQSGSV 1375
Query: 286 GMILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIG-ISTTDTIKSVIKIRMSQPK 344
+ P K+ ++ +L IN + H +G I T TI
Sbjct: 1376 AFYYVDFPVTEVKSKHGDA-ILQYIN-------STSHPVGTILATVTIPDY--------- 1504
Query: 345 TSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFP 404
KPAP + FSSRGP+ + +LKPD+ APGVNILAA+ F + ++ V R+
Sbjct: 1505 -----KPAPRVGYFSSRGPSLITSNVLKPDIAAPGVNILAAW--FGNDTSEVPKGRKPSL 1663
Query: 405 FNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLA 464
+ I GTSM+ PHV+G A +K +P WS +AIKSAIMT+A DN I +A
Sbjct: 1664 YRILSGTSMATPHVSGLACSVKRKNPTWSASAIKSAIMTSAIQNDNLKGPI-TTDSGLIA 1840
Query: 465 NPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNP-NMTFTC---S 520
P+ YG+G I + + PGLVY+ + VDYLN+LC G + +I + F C S
Sbjct: 1841 TPYDYGAGAITTSEPLQPGLVYETNNVDYLNYLCYNGLNITMIKVISGTVPENFNCPKDS 2020
Query: 521 GIHSINDLNYPSITLPNLGLNAVNVTRIVTNVG--PPSTYFAKVQLPGYNIVVV-PDSLT 577
I+ +NYPSI + G V+R VTNV + YF V+ P IV + P +L
Sbjct: 2021 SSDLISSINYPSIAVNFTGKADAVVSRTVTNVDEEDETVYFPVVEAPSEVIVTLFPYNLE 2200
Query: 578 FKKNGEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTVQR 624
F + +K+ + + R T + FG + W+N K++VR P + +
Sbjct: 2201 FTTSIKKQSYNITF--RPKTSLKKDLFGSITWSNDKYMVRIPFVLTK 2335
>TC226350 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(34%)
Length = 1312
Score = 257 bits (657), Expect = 9e-69
Identities = 150/344 (43%), Positives = 202/344 (58%), Gaps = 2/344 (0%)
Frame = +1
Query: 281 SAGAKGMILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIKSVIKIRM 340
SAGA GM+L N NG+ L+A++++L K G +I K +KI
Sbjct: 7 SAGALGMVLSNTAA-NGEELVADAHLLPATAVGQK----AGDAIKKYLFSDAKPTVKILF 171
Query: 341 SQPKTSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNR 400
K + P+PV+A+FSSRGPN + P ILKPD+ APGVNILA +S + L DNR
Sbjct: 172 EGTKLGIQ--PSPVVAAFSSRGPNSITPQILKPDLIAPGVNILAGWSKAVGPTGLPVDNR 345
Query: 401 RGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAID 460
R FNI GTSMSCPHV+G A LIK+ HP+WSPAA++SA+MTTA T + ++D+
Sbjct: 346 R-VDFNIISGTSMSCPHVSGLAALIKSAHPDWSPAAVRSALMTTAYTVYKTGEKLQDSAT 522
Query: 461 KTLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCS 520
+ PF +GSGH+ P A++PGLVYDL+V DYL FLCA YS I+TL
Sbjct: 523 GKPSTPFDHGSGHVDPVAALNPGLVYDLTVDDYLGFLCALNYSASEINTLAKRKFQCDAG 702
Query: 521 GIHSINDLNYPSITLPNLGLNAVNVTRIVTNVGPPSTYFAKV--QLPGYNIVVVPDSLTF 578
+S+ DLNYPS + V TR +TNVGP TY A V + I V P L+F
Sbjct: 703 KQYSVTDLNYPSFAVLFESGGVVKHTRTLTNVGPAGTYKASVTSDMASVKISVEPQVLSF 882
Query: 579 KKNGEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTV 622
K+N EKK F V + + FG ++W++GKH+V +P+++
Sbjct: 883 KEN-EKKSFTVTFSSSGSPQQRVNAFGRVEWSDGKHVVGTPISI 1011
>TC205344 similar to UP|Q9SZV5 (Q9SZV5) Proteinase-like protein (AT4g30020
protein) (AT4g30020/F6G3_50), partial (78%)
Length = 2192
Score = 254 bits (649), Expect = 8e-68
Identities = 209/615 (33%), Positives = 303/615 (48%), Gaps = 31/615 (5%)
Frame = +2
Query: 37 NGVGEVQRKFHVTAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVC 96
+G G + AG N +P + G G G +P++R+ YK + + G
Sbjct: 221 DGDGHGSHTASIAAGRNGIP-VRMHGHEFGKASGMAPRARIAVYKALYR--LFGG----- 376
Query: 97 YGADVLSAIDQAISDGVDIISVSVGGRSS-SNFEEIFTDEISIGAFQAFAKNILLVASAG 155
+ ADV++AIDQA+ DGVDI+S+SVG S SN + F + A + + +AG
Sbjct: 377 FIADVVAAIDQAVHDGVDILSLSVGPNSPPSNTKTTFLNPFDATLLGAVKAGVFVAQAAG 556
Query: 156 NGGPTPGSVTNVAPWVFTVAASTIDRDFSSTITIGN-KTVTGASLFVNLPPNQSFTLVDS 214
NGGP P S+ + +PW+ TVAA+ DR + + + +GN K + G L + NQ++TLV +
Sbjct: 557 NGGPFPKSLVSYSPWIATVAAAIDDRRYKNHLILGNGKILAGLGLSPSTRLNQTYTLVAA 736
Query: 215 IDAKF-ANVTNQDARFC-KPGTLDPSKVSGKIVEC------VGEKITIKNTSEPVSGR-L 265
D ++VT C +P L+ + + G I+ C V +IK SE
Sbjct: 737 TDVLLDSSVTKYSPTDCQRPELLNKNLIKGNILLCGYSYNFVIGSASIKQVSETAKALGA 916
Query: 266 LGFA--TNSVSQGR--EALSAGAKGMILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRG 321
+GF +VS G + + G G+++ + K K L+ N+ + ++ + + G
Sbjct: 917 VGFVLYVENVSPGTKFDPVPVGIPGILITDASK--SKELIDYYNISTPRDWTGRVKTFEG 1090
Query: 322 HSIGISTTDTIKSVIKIRMSQPKTSYRRKPAPVMASFSSRGPNQV-----QPYILKPDVT 376
D + ++ K AP +A FS+RGPN + +LKPD+
Sbjct: 1091TG---KIEDGLMPIL------------HKSAPQVAMFSARGPNIKDFSFQEADLLKPDIL 1225
Query: 377 APGVNILAAYSLFASVSNLVTDNRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAA 436
APG I AA+SL + N G F + GTSM+ PH+AG A LIK HP+WSPAA
Sbjct: 1226APGSLIWAAWSL----NGTDEPNYAGEGFAMISGTSMAAPHIAGIAALIKQKHPHWSPAA 1393
Query: 437 IKSAIMTTATIRDNTNKLI-------RDAIDKTLANPFAYGSGHIQPNTAMDPGLVYDLS 489
IKSA+MTT+T D I +A+ A PF YGSGH+ P A+DPGL++D
Sbjct: 1394IKSALMTTSTTLDRAGNPILAQLYSETEAMKLVKATPFDYGSGHVNPRAALDPGLIFDAG 1573
Query: 490 VVDYLNFLCAA-GYSQRLISTLLNPNMTFTCSGIHSINDLNYPSITLPNLGLNAVNVTRI 548
DYL FLC G I N T H N LN PSIT+ +L + + VTR
Sbjct: 1574YEDYLGFLCTTPGIDVHEIKNYTNSPCNNTMG--HPSN-LNTPSITISHLVRSQI-VTRT 1741
Query: 549 VTNVG-PPSTYFAKVQL-PGYNIVVVPDSLTFKKNGEKKKFQVIVQARSVTPRGRYQFGE 606
VTNV TY ++ P I V P ++T K + ++F V + RSVT G Y FGE
Sbjct: 1742VTNVADEEETYVITARMQPAVAIDVNPPAMTIKASA-SRRFTVTLTVRSVT--GTYSFGE 1912
Query: 607 LQWTNGK-HIVRSPV 620
+ + H VR PV
Sbjct: 1913VLMKGSRGHKVRIPV 1957
>TC229882 weakly similar to UP|Q75I27 (Q75I27) Putaive subtilisin-like
proteinase, partial (27%)
Length = 1263
Score = 230 bits (587), Expect = 1e-60
Identities = 124/274 (45%), Positives = 162/274 (58%), Gaps = 3/274 (1%)
Frame = +1
Query: 350 KPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQ 409
KP+PV+A+FSSRGPN V ILKPDV PGVNILA +S S L D R+ FNI
Sbjct: 160 KPSPVVAAFSSRGPNMVTRQILKPDVIGPGVNILAGWSEAIGPSGLSDDTRKT-QFNIMS 336
Query: 410 GTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPFAY 469
GTSMSCPH++G A L+K HP WS +AIKSA+MTTA + DNT +RDA +NP+A+
Sbjct: 337 GTSMSCPHISGLAALLKAAHPQWSSSAIKSALMTTADVHDNTKSQLRDAAGGAFSNPWAH 516
Query: 470 GSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIHSINDLN 529
G+GH+ P+ A+ PGLVYD + DY+ FLC+ Y+ I + + LN
Sbjct: 517 GAGHVNPHKALSPGLVYDATPSDYIKFLCSLEYTPERIQLITKRSGVNCTKRFSDPGQLN 696
Query: 530 YPSITLPNLGLNAVNVTRIVTNVGPP-STYFAKVQLPG-YNIVVVPDSLTFKKNGEKKKF 587
YPS ++ G V TR++TNVG S Y V P + V P +L F K GE++++
Sbjct: 697 YPSFSVLFGGKRVVRYTRVLTNVGEAGSVYNVTVDAPSTVTVTVKPAALVFGKVGERQRY 876
Query: 588 -QVIVQARSVTPRGRYQFGELQWTNGKHIVRSPV 620
V V RY FG + W+N +H VRSPV
Sbjct: 877 TATFVSKNGVGDSVRYGFGSIMWSNAQHQVRSPV 978
>TC229906 weakly similar to UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C1,
partial (40%)
Length = 1392
Score = 225 bits (574), Expect = 4e-59
Identities = 175/475 (36%), Positives = 238/475 (49%), Gaps = 10/475 (2%)
Frame = +2
Query: 158 GPTPGSVTNVAPWVFTVAASTIDRDFSSTITIGNKTV-TGASLFVNLPPNQSFTLVDSID 216
GP S+T +PW+ +VAASTI R F + + +GN V G S+ N+ F LV + D
Sbjct: 5 GPGLSSITTYSPWILSVAASTIGRKFLTKVQLGNGMVFEGVSINTFDLKNKMFPLVYAGD 184
Query: 217 AKFA--NVTNQDARFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVS 274
+ +RFC ++D V GKIV C G N S G L
Sbjct: 185 VPNTADGYNSSTSRFCYVNSVDKHLVKGKIVLCDG------NASPKKVGDL--------- 319
Query: 275 QGREALSAGAKGMIL-RNQPKFNGKTLLAESNVLSTINYYDKHQLTRGHSIGISTTDTIK 333
+GA GM+L K T + +S N+ HS +S ++
Sbjct: 320 -------SGAAGMLLGATDVKDAPFTYALPTAFISLRNF------KLIHSYMVSLRNSTA 460
Query: 334 SVIKIRMSQPKTSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVS 393
++ + + P + SFSSRGPN + P LKPD+ APGVNILAA+S ++S
Sbjct: 461 TIFRSDEDNDDSQ-----TPFIVSFSSRGPNPLTPNTLKPDLAAPGVNILAAWSPVYTIS 625
Query: 394 NLVTDNRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNK 453
D +R +NI+ GTSM+CPHV+ A +K+ HPNWSPA IKSA+MTTAT T
Sbjct: 626 EFKGD-KRAVQYNIESGTSMACPHVSAAAAYVKSFHPNWSPAMIKSALMTTATPMSPT-- 796
Query: 454 LIRDAIDKTLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNP 513
L DA FAYG+G I P A +PGLVYD+S DY+ FLC GY+ ++ L
Sbjct: 797 LNPDA-------EFAYGAGLINPLKAANPGLVYDISEADYVKFLCGEGYTDEMLRVLTKD 955
Query: 514 NMTFTCS---GIHSINDLNYPSITL-PNLGLNAVNVTRIVTNVG-PPSTYFAKVQLPG-Y 567
+ CS ++ DLN PS+ L N+ + R VTNVG S+Y AKV P
Sbjct: 956 HS--RCSKHAKKEAVYDLNLPSLALYVNVSSFSRIFHRTVTNVGLATSSYKAKVVSPSLI 1129
Query: 568 NIVVVPDSLTFKKNGEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTV 622
+I V P+ L+F G+KK F VI++ +V P L W +G VRSP+ V
Sbjct: 1130DIQVKPNVLSFTSIGQKKSFSVIIEG-NVNP--DILSASLVWDDGTFQVRSPIVV 1285
>TC207663 similar to UP|P93204 (P93204) SBT1 protein (Subtilisin-like
protease) , partial (37%)
Length = 1171
Score = 225 bits (573), Expect = 5e-59
Identities = 128/289 (44%), Positives = 166/289 (57%), Gaps = 13/289 (4%)
Frame = +1
Query: 345 TSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFP 404
T + +P+PV+A+FSSRGPN + P ILKPD+ APGVNILA ++ + L D R
Sbjct: 58 THLQVQPSPVVAAFSSRGPNALTPKILKPDLIAPGVNILAGWTGAVGPTGLTVDTRH-VS 234
Query: 405 FNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLA 464
FNI GTSMSCPHV+G A ++K HP WSPAAI+SA+MTTA + I+D
Sbjct: 235 FNIISGTSMSCPHVSGLAAILKGAHPQWSPAAIRSALMTTAYTSYKNGETIQDISTGQPG 414
Query: 465 NPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIHS 524
PF YG+GH+ P A+DPGLVYD +V DYL F CA YS I + T +
Sbjct: 415 TPFDYGAGHVDPVAALDPGLVYDANVDDYLGFFCALNYSSFQIKLAARRDYTCDPKKDYR 594
Query: 525 INDLNYPSITLP-----NLG-----LNAVNVTRIVTNVGPPSTYFAKVQLPG---YNIVV 571
+ D NYPS +P +G L V +R++TNVG P TY A V G VV
Sbjct: 595 VEDFNYPSFAVPMDTASGIGGGSDTLKTVKYSRVLTNVGAPGTYKASVMSLGDSNVKTVV 774
Query: 572 VPDSLTFKKNGEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPV 620
P++L+F + EKK + V S+ P G F L+WT+GKH V SP+
Sbjct: 775 EPNTLSFTELYEKKDYTVSFTYTSM-PSGTTSFARLEWTDGKHKVGSPI 918
>TC216428 similar to UP|Q9AX30 (Q9AX30) Subtilisin-like protease, partial
(12%)
Length = 810
Score = 224 bits (572), Expect = 7e-59
Identities = 115/198 (58%), Positives = 137/198 (69%), Gaps = 1/198 (0%)
Frame = +1
Query: 426 KTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPFAYGSGHIQPNTAMDPGLV 485
KT HP WSPAAIKSAIMTTAT DNTN+ IR+A K +A PF YG+GHIQPN A+DPGLV
Sbjct: 1 KTYHPTWSPAAIKSAIMTTATTLDNTNQPIRNAFHK-VATPFEYGAGHIQPNLAIDPGLV 177
Query: 486 YDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIHSINDLNYPSITLPNLGLNAVNV 545
YDL DYLNFLCA+GY+Q L++ +TC + I D NYPSIT+ + G ++V
Sbjct: 178 YDLRTTDYLNFLCASGYNQALLNLFAKLKFPYTCPKSYRIEDFNYPSITVRHPGSKTISV 357
Query: 546 TRIVTNVGPPSTYFAKVQLP-GYNIVVVPDSLTFKKNGEKKKFQVIVQARSVTPRGRYQF 604
TR VTNVGPPSTY P G ++V P SLTFK+ GEKKKFQVI+Q R F
Sbjct: 358 TRTVTNVGPPSTYVVNTHGPKGIKVLVQPSSLTFKRTGEKKKFQVILQPIGAR---RGLF 528
Query: 605 GELQWTNGKHIVRSPVTV 622
G L WT+GKH V SP+T+
Sbjct: 529 GNLSWTDGKHRVTSPITI 582
>AW234015
Length = 411
Score = 223 bits (567), Expect = 3e-58
Identities = 109/136 (80%), Positives = 119/136 (87%)
Frame = +3
Query: 450 NTNKLIRDAIDKTLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLIST 509
NTNK I DA DKTLANPFAYGSGH+QPN+A+DPGL+YDLS+VDYLNFLCA+GY Q+LIS
Sbjct: 3 NTNKPIGDAFDKTLANPFAYGSGHVQPNSAIDPGLIYDLSIVDYLNFLCASGYDQQLISA 182
Query: 510 LLNPNMTFTCSGIHSINDLNYPSITLPNLGLNAVNVTRIVTNVGPPSTYFAKVQLPGYNI 569
L N N TFTCSG HSI DLNYPSITLPNLGLNA+ VTR VTNVGP STYFAK QL GYNI
Sbjct: 183 L-NFNSTFTCSGSHSITDLNYPSITLPNLGLNAITVTRTVTNVGPASTYFAKAQLRGYNI 359
Query: 570 VVVPDSLTFKKNGEKK 585
VVVP SL+FKK GEK+
Sbjct: 360 VVVPSSLSFKKIGEKR 407
>TC228967 similar to GB|BAB09208.1|9758669|AB012245 subtilisin-like protease
{Arabidopsis thaliana;} , partial (21%)
Length = 1106
Score = 215 bits (547), Expect = 5e-56
Identities = 133/286 (46%), Positives = 164/286 (56%), Gaps = 8/286 (2%)
Frame = +2
Query: 345 TSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFP 404
T KPAP MASFSSRGPN V P ILKPD+TAPGV+ILAA++ + + +++R
Sbjct: 50 TVLETKPAPSMASFSSRGPNIVDPNILKPDITAPGVDILAAWTAEDGPTRMTFNDKRVVK 229
Query: 405 FNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLA 464
+NI GTSMSCPHVA A L+K +HP WS AAI+SA+MTTA DNT + D A
Sbjct: 230 YNIFSGTSMSCPHVAAAAVLLKAIHPTWSTAAIRSALMTTAMTTDNTGHPLTDETGNP-A 406
Query: 465 NPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTC-SGIH 523
PFA GSGH P A DPGLVYD S + YL + C G +Q N N+T+ C
Sbjct: 407 TPFAMGSGHFNPKRAADPGLVYDASYMGYLLYTCNLGVTQ-------NFNITYNCPKSFL 565
Query: 524 SINDLNYPSITLPNLGLNAVNVTRIVTNVG-PPSTY-FAKVQLPGYNIVVVPDSLTFKKN 581
+LNYPSI + L + R VTNVG S Y F+ V Y+I P+ L F
Sbjct: 566 EPFELNYPSIQIHRL-YYTKTIKRTVTNVGRGRSVYKFSAVSPKEYSITATPNILKFNHV 742
Query: 582 GEKKKFQVIVQA---RSVTPRG--RYQFGELQWTNGKHIVRSPVTV 622
G+K F + V A + T G +Y FG WT+ HIVRSPV V
Sbjct: 743 GQKINFTITVTANWSQIPTKHGPDKYYFGWYAWTHQHHIVRSPVAV 880
>TC233666 weakly similar to UP|Q9FJF3 (Q9FJF3) Serine protease-like protein,
partial (30%)
Length = 678
Score = 203 bits (516), Expect = 2e-52
Identities = 113/244 (46%), Positives = 149/244 (60%), Gaps = 1/244 (0%)
Frame = +1
Query: 49 TAGGNFVPGASIFGIGNGTIKGGSPKSRVVTYKVCWSQTIADGNSAVCYGADVLSAIDQA 108
T GG FVPGA++FG+GNGT +GGSP++RV TYKVCW DGN C+ AD+++A D A
Sbjct: 22 TIGGTFVPGANVFGLGNGTAEGGSPRARVATYKVCWPPI--DGNE--CFDADIMAAFDMA 189
Query: 109 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 168
I DGVD++S+S+GG N + F D +SIGAF A K I ++ SAGN GPTP +V NVA
Sbjct: 190 IHDGVDVLSLSLGG----NATDYFDDGLSIGAFHANMKGIPVICSAGNYGPTPATVFNVA 357
Query: 169 PWVFTVAASTIDRDFSSTITIGN-KTVTGASLFVNLPPNQSFTLVDSIDAKFANVTNQDA 227
PW+ TV AST+DR F S + + N + GASL +P ++ + L+++ DAK AN ++A
Sbjct: 358 PWILTVGASTLDRQFDSVVELHNGQRFMGASLSKAMPEDKLYPLINAADAKAANKPVENA 537
Query: 228 RFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKGM 287
C GT+DP K GKI+ C L T V + AL AGA M
Sbjct: 538 TLCMRGTIDPEKARGKILVC------------------LRGVTARVEKSLVALEAGAAXM 663
Query: 288 ILRN 291
IL N
Sbjct: 664 ILCN 675
>TC207701 similar to UP|Q84TR6 (Q84TR6) Subtilase, partial (6%)
Length = 763
Score = 202 bits (515), Expect = 3e-52
Identities = 100/150 (66%), Positives = 119/150 (78%), Gaps = 1/150 (0%)
Frame = +2
Query: 476 PNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTCSGIHSINDLNYPSITL 535
P+ A++PGLVYDLS+ DYLNFLCA+GY Q+LIS L N N TF CSG HS+NDLNYPSITL
Sbjct: 2 PDLAIEPGLVYDLSLTDYLNFLCASGYDQQLISAL-NFNRTFICSGSHSVNDLNYPSITL 178
Query: 536 PNLGLNAVNVTRIVTNVGPPSTYFAKVQLP-GYNIVVVPDSLTFKKNGEKKKFQVIVQAR 594
PNL L V + R VTNVGPPSTY + P GY+I VVP SLTF K GE+K F+VIVQA
Sbjct: 179 PNLRLKPVTIARTVTNVGPPSTYTVSTRSPNGYSIAVVPPSLTFTKIGERKTFKVIVQAS 358
Query: 595 SVTPRGRYQFGELQWTNGKHIVRSPVTVQR 624
S R +Y+FG+L+WT+GKHIVRSP+TV+R
Sbjct: 359 SAATRRKYEFGDLRWTDGKHIVRSPITVKR 448
>TC229044 similar to UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease, partial
(42%)
Length = 984
Score = 202 bits (515), Expect = 3e-52
Identities = 121/284 (42%), Positives = 175/284 (61%), Gaps = 9/284 (3%)
Frame = +1
Query: 350 KPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFNIQQ 409
KPAPV+ +FSSRGP+ + ILKPD+ APGVNILAA+ + ++ V R+ +NI
Sbjct: 124 KPAPVVPNFSSRGPSSLSSNILKPDIAAPGVNILAAW--IGNNADDVPKGRKPSLYNIIS 297
Query: 410 GTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNTNKLIRDAIDKTLANPFAY 469
GTSM+CPHV+G A +KT +P WS +AIKSAIMT+A I+ N K +A P+ Y
Sbjct: 298 GTSMACPHVSGLASSVKTRNPTWSASAIKSAIMTSA-IQINNLKAPITTDSGRVATPYDY 474
Query: 470 GSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGY---SQRLISTLLNPNMTFTC---SGIH 523
G+G + + ++ PGLVY+ + +DYLN+LC G + ++IS + N F+C S
Sbjct: 475 GAGEMTTSESLQPGLVYETNTIDYLNYLCYIGLNITTVKVISRTVPAN--FSCPKDSSSD 648
Query: 524 SINDLNYPSITLPNLGLNAVNVTRIVTNVG--PPSTYFAKVQLP-GYNIVVVPDSLTFKK 580
I+++NYPSI + G AVNV+R VTNVG + Y V+ P G + V PD L F K
Sbjct: 649 LISNINYPSIAVNFTGKAAVNVSRTVTNVGEEDETAYSPVVEAPSGVKVTVTPDKLQFTK 828
Query: 581 NGEKKKFQVIVQARSVTPRGRYQFGELQWTNGKHIVRSPVTVQR 624
+ +K +QVI + ++T FG + W+NGK++VRSP + +
Sbjct: 829 SSKKLGYQVIFSS-TLTSLKEDLFGSITWSNGKYMVRSPFVLTK 957
>TC205345 similar to UP|Q9SZV5 (Q9SZV5) Proteinase-like protein (AT4g30020
protein) (AT4g30020/F6G3_50), partial (62%)
Length = 1729
Score = 201 bits (510), Expect = 1e-51
Identities = 173/523 (33%), Positives = 257/523 (49%), Gaps = 36/523 (6%)
Frame = +1
Query: 134 DEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVAPWVFTVAASTIDRDFSSTITIGN-K 192
D +GA +A + + +AGN GP P ++ + +PW+ +VAA+ DR + + + +GN K
Sbjct: 40 DATLLGAVKA---GVFVAQAAGNHGPLPKTLVSYSPWIASVAAAIDDRRYKNHLILGNGK 210
Query: 193 TVTGASLFVNLPPNQSFTLVDS----IDAKFANVTNQDARFCKPGTLDPSKVSGKIVECV 248
T+ G L + N+++TLV + +D+ + D + +P L+ + + G I+ C
Sbjct: 211 TLAGIGLSPSTHLNETYTLVAANDVLLDSSLMKYSPTDCQ--RPELLNKNLIKGNILLCG 384
Query: 249 GEKITIKNTSEPVSGRLLGFATNSVSQGREALSAGAKGMILRNQ-----PKFN------- 296
+ T+ + VS+ +AL GA G +L + KFN
Sbjct: 385 YSFNFVVGTA----------SIKKVSETAKAL--GAVGFVLCVENISLGTKFNPVPVGLP 528
Query: 297 GKTLLAESNVLSTINYYDKHQLTRGHSIGIST----TDTIKSVI-KIRMSQPKTSYRRKP 351
G ++ SN I+YY+ I+T T +KS K ++ K
Sbjct: 529 GILIIDVSNSKELIDYYN-----------ITTPRDWTGRVKSFEGKGKIGDGLMPILHKS 675
Query: 352 APVMASFSSRGPNQV-----QPYILKPDVTAPGVNILAAYSLFASVSNLVTDNRRGFPFN 406
AP +A FS+RGPN + +LKPD+ APG I AA+ + N G F
Sbjct: 676 APQVALFSARGPNIKDFSFQEADLLKPDILAPGSLIWAAWC----PNGTDEPNYVGEAFA 843
Query: 407 IQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTATIRDNT-NKLI------RDAI 459
+ GTSM+ PH+AG A LIK HP+WSPAAIKSA+MTT+T D N L+ +A+
Sbjct: 844 MISGTSMAAPHIAGIAALIKQKHPHWSPAAIKSALMTTSTTLDRAGNPLLAQQTSESEAM 1023
Query: 460 DKTLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGYSQRLISTLLNPNMTFTC 519
A PF YGSGH+ P A+DPGL++D DY+ FLC + + + T
Sbjct: 1024RLVKATPFDYGSGHVDPTAALDPGLIFDAGYKDYVGFLCTTPSID--VHEIRHYTHTPCN 1197
Query: 520 SGIHSINDLNYPSITLPNLGLNAVNVTRIVTNVGPPSTYFAKVQL-PGYNIVVVPDSLTF 578
+ + ++LN PSIT+ L V VTR VTNV TY ++ P I V P ++T
Sbjct: 1198TTMGKPSNLNTPSITISYLVRTQV-VTRTVTNVAEEETYVITARMEPAVAIEVNPPAMTI 1374
Query: 579 KKNGEKKKFQVIVQARSVTPRGRYQFGELQWTNGK-HIVRSPV 620
K G ++F V + RSVT RY FGE+ + H VR PV
Sbjct: 1375KA-GASRQFSVSLTVRSVT--RRYSFGEVLMKGSRGHKVRIPV 1494
>TC221491 similar to UP|P93204 (P93204) SBT1 protein (Subtilisin-like
protease) , partial (34%)
Length = 872
Score = 196 bits (499), Expect = 2e-50
Identities = 130/318 (40%), Positives = 162/318 (50%)
Frame = +3
Query: 203 LPPNQSFTLVDSIDAKFANVTNQDARFCKPGTLDPSKVSGKIVECVGEKITIKNTSEPVS 262
LPPN +V + ANV+++ C GTL KV+GKIV C
Sbjct: 15 LPPNSPLPIVYA-----ANVSDESQNLCTRGTLIAEKVAGKIVICDRG------------ 143
Query: 263 GRLLGFATNSVSQGREALSAGAKGMILRNQPKFNGKTLLAESNVLSTINYYDKHQLTRGH 322
V +G SAG GMIL N + G+ L+A+S +L K
Sbjct: 144 ------GNARVEKGLVVKSAGGIGMILSNNEDY-GEELVADSYLLPAAALGQKSSNELKK 302
Query: 323 SIGISTTDTIKSVIKIRMSQPKTSYRRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNI 382
+ S T K + T +P+PV+A+FSSRGPN + P ILKPD+ APGVNI
Sbjct: 303 YVFSSPNPTAK------LGFGGTQLGVQPSPVVAAFSSRGPNVLTPKILKPDLIAPGVNI 464
Query: 383 LAAYSLFASVSNLVTDNRRGFPFNIQQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIM 442
LA ++ + L D R FNI GTSMSCPHV G A L+K HP WSPAAI+SA+M
Sbjct: 465 LAGWTGAVGPTGLTEDTRH-VEFNIISGTSMSCPHVTGLAALLKGTHPEWSPAAIRSALM 641
Query: 443 TTATIRDNTNKLIRDAIDKTLANPFAYGSGHIQPNTAMDPGLVYDLSVVDYLNFLCAAGY 502
TTA + I+D A PF YG+GH+ P A DPGLVYD +V DYL+F CA Y
Sbjct: 642 TTAYRTYKNGQTIKDVATGLPATPFDYGAGHVDPVAAFDPGLVYDTTVDDYLSFFCALNY 821
Query: 503 SQRLISTLLNPNMTFTCS 520
S I L FTCS
Sbjct: 822 SPYQIK--LVARRDFTCS 869
>BE021282 similar to GP|21593457|gb| subtilisin-like serine protease
{Arabidopsis thaliana}, partial (5%)
Length = 402
Score = 185 bits (470), Expect = 5e-47
Identities = 90/132 (68%), Positives = 107/132 (80%)
Frame = +1
Query: 109 ISDGVDIISVSVGGRSSSNFEEIFTDEISIGAFQAFAKNILLVASAGNGGPTPGSVTNVA 168
I DGVDIIS+S GG E IFTDE+SIGAF A A+N +LVASAGN GPTPG+V NVA
Sbjct: 1 IDDGVDIISLSAGGSYVVTPEGIFTDEVSIGAFHAIARNRILVASAGNDGPTPGTVLNVA 180
Query: 169 PWVFTVAASTIDRDFSSTITIGNKTVTGASLFVNLPPNQSFTLVDSIDAKFANVTNQDAR 228
PWVFT+AAST+DRDFSS +TI N+ +TGASLFVNLPPN++F+L+ + DAK AN T +DA
Sbjct: 181 PWVFTIAASTLDRDFSSNLTINNRQITGASLFVNLPPNKAFSLILATDAKLANATFRDAE 360
Query: 229 FCKPGTLDPSKV 240
C+PGTLDP KV
Sbjct: 361 LCRPGTLDPEKV 396
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,330,406
Number of Sequences: 63676
Number of extensions: 289376
Number of successful extensions: 1687
Number of sequences better than 10.0: 118
Number of HSP's better than 10.0 without gapping: 1513
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1527
length of query: 624
length of database: 12,639,632
effective HSP length: 103
effective length of query: 521
effective length of database: 6,081,004
effective search space: 3168203084
effective search space used: 3168203084
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC149129.14


