

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149127.4 + phase: 0
(323 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
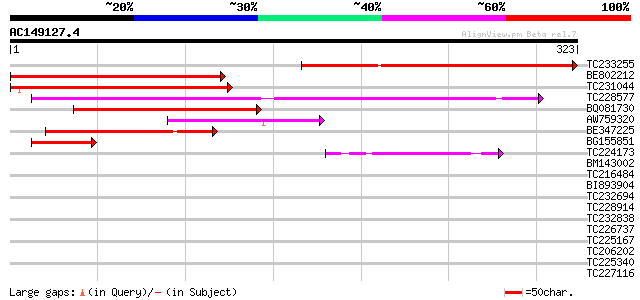
Score E
Sequences producing significant alignments: (bits) Value
TC233255 similar to UP|Q6NMC6 (Q6NMC6) At5g24310, partial (25%) 220 6e-58
BE802212 206 1e-53
TC231044 similar to UP|Q6NMC6 (Q6NMC6) At5g24310, partial (38%) 203 1e-52
TC228577 146 1e-35
BQ081730 weakly similar to GP|20197375|gb expressed protein {Ara... 126 1e-29
AW759320 118 3e-27
BE347225 74 7e-14
BG155851 similar to GP|20197375|gb| expressed protein {Arabidops... 42 3e-04
TC224173 similar to UP|O81330 (O81330) F3D13.1 protein, partial ... 41 8e-04
BM143002 40 0.001
TC216484 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabin... 38 0.005
BI893904 similar to GP|15809850|gb AT5g37540/mpa22_p_70 {Arabido... 38 0.007
TC232694 38 0.007
TC228914 similar to UP|DAH1_MOUSE (Q9QYB2) Dachshund homolog 1 (... 36 0.021
TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {A... 36 0.027
TC226737 similar to UP|Q9SEE9 (Q9SEE9) Arginine/serine-rich prot... 36 0.027
TC225167 similar to GB|CAA44226.1|20020|NTRPL121A ribosomal prot... 35 0.035
TC206202 UP|Q99367 (Q99367) DNA-directed RNA polymerase (Fragme... 35 0.035
TC225340 similar to UP|Q9SPI1 (Q9SPI1) Splicing factor SR1, part... 35 0.046
TC227116 similar to UP|WAS2_HUMAN (Q9Y6W5) Wiskott-Aldrich syndr... 35 0.060
>TC233255 similar to UP|Q6NMC6 (Q6NMC6) At5g24310, partial (25%)
Length = 666
Score = 220 bits (561), Expect = 6e-58
Identities = 115/158 (72%), Positives = 132/158 (82%), Gaps = 1/158 (0%)
Frame = +1
Query: 167 DDWHHFRNAVRATIRETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPH 226
D+W +FRNAVRATIRETP ST+ +G SPSPSLQPQR FSFTS M KKDLE+R+VSP+
Sbjct: 1 DEWPNFRNAVRATIRETPPSTARRGRSPSPSLQPQRSRDFSFTS-TMPKKDLERRSVSPY 177
Query: 227 RFPLSRTGSMSS-RSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSSDVN 285
RFPL RTGS SS R TTPK R TTPN S T +PSNAR RYPSEPRKS+SMRLS++ +
Sbjct: 178 RFPLIRTGSRSSSRPTTPKISRPTTPNPSRPTTPNPSNARQRYPSEPRKSSSMRLSAERD 357
Query: 286 NIRDIDQHPSKSKRLLKSLLSRRKSKKDDTLYTYLDEY 323
+ +DI+Q+PSKSKRLLK+LLSRRKSKKDD LYTYLDEY
Sbjct: 358 HGKDIEQYPSKSKRLLKALLSRRKSKKDDMLYTYLDEY 471
>BE802212
Length = 411
Score = 206 bits (524), Expect = 1e-53
Identities = 101/123 (82%), Positives = 109/123 (88%)
Frame = +3
Query: 1 MGKIATQPLPRMASNYDEVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQI 60
MGKIATQPL R ASNYDEVFM Q+LLFDDSL DLKNLRTQLYSAAEYFELSY NDDQKQI
Sbjct: 39 MGKIATQPLSREASNYDEVFMQQSLLFDDSLKDLKNLRTQLYSAAEYFELSYANDDQKQI 218
Query: 61 LVETLKDYAVKALINTVDHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQGFMD 120
++ETLKDYA+KALIN+VDHLGSV YKV+DLLDEK+ EV LRLSCI+QRI TC FMD
Sbjct: 219 VIETLKDYAIKALINSVDHLGSVTYKVNDLLDEKIVEVSETQLRLSCIQQRISTCHAFMD 398
Query: 121 HEG 123
HEG
Sbjct: 399 HEG 407
>TC231044 similar to UP|Q6NMC6 (Q6NMC6) At5g24310, partial (38%)
Length = 538
Score = 203 bits (516), Expect = 1e-52
Identities = 101/130 (77%), Positives = 114/130 (87%), Gaps = 3/130 (2%)
Frame = +1
Query: 1 MGKI---ATQPLPRMASNYDEVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQ 57
MGK+ ATQP+ + ASNYDEVFM Q+LLFDDSL DLKNLR QLYSAAEYFELSY+NDDQ
Sbjct: 148 MGKVTTSATQPVSQEASNYDEVFMQQSLLFDDSLKDLKNLRAQLYSAAEYFELSYSNDDQ 327
Query: 58 KQILVETLKDYAVKALINTVDHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQG 117
KQ++VETLKDYA+KAL+NTVDHLGSV YKV+DLLDEKV EV +LR+SCIEQRIKTC
Sbjct: 328 KQVVVETLKDYAIKALVNTVDHLGSVTYKVNDLLDEKVGEVSVAELRVSCIEQRIKTCHE 507
Query: 118 FMDHEGHTQQ 127
+MDHEG TQQ
Sbjct: 508 YMDHEGRTQQ 537
>TC228577
Length = 1071
Score = 146 bits (368), Expect = 1e-35
Identities = 97/293 (33%), Positives = 151/293 (51%), Gaps = 1/293 (0%)
Frame = +3
Query: 13 ASNYDEVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQILVETLKDYAVKA 72
A YDE M ++ F ++L +LKNLR QLYSAAEY E SY + +QKQ++++ LKDYAV+A
Sbjct: 216 AMTYDEASMERSKSFVNALQELKNLRPQLYSAAEYCEKSYLHSEQKQMVLDNLKDYAVRA 395
Query: 73 LINTVDHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQRIKTCQGFMDHEGHTQQSLVIS 132
L+N VDHLG+VAYK++DLL+++ +V DL+++ I Q++ TCQ + D EG QQ L+
Sbjct: 396 LVNAVDHLGTVAYKLTDLLEQQTFDVSTMDLKVATINQKLLTCQIYTDKEGLRQQQLLAF 575
Query: 133 TPKHHKRYILPVGETLHGTNSTKSKYIGCHLDDEDDWHHFRNAVRATIRETPTSTSSKGN 192
P+HHK YILP + K + + + + F+ R TP + + +
Sbjct: 576 IPRHHKHYILP-------NSVNKKVHFSPQIQIDARQNPFQTRTRFQSSGTPAAKTLSWH 734
Query: 193 SPSPSLQPQRVGAFSFTSPNMAK-KDLEKRTVSPHRFPLSRTGSMSSRSTTPKTGRSTTP 251
S + + + + SPN+ K K + H M S +
Sbjct: 735 LASETKSTLKGSSHASPSPNIENPKFSGKASGVFHLLDNEENTWMKSSPAQIHFPNGVST 914
Query: 252 NSSNRATTSPSNARVRYPSEPRKSASMRLSSDVNNIRDIDQHPSKSKRLLKSL 304
+S+ T + S+P + S D N R+ Q P++SK +L +L
Sbjct: 915 SSTPMHTLGGTRKDALEGSKPFTTFR---SFDNQNRRETVQVPTRSKSMLSAL 1064
>BQ081730 weakly similar to GP|20197375|gb expressed protein {Arabidopsis
thaliana}, partial (45%)
Length = 422
Score = 126 bits (317), Expect = 1e-29
Identities = 59/107 (55%), Positives = 80/107 (74%)
Frame = +1
Query: 37 LRTQLYSAAEYFELSYTNDDQKQILVETLKDYAVKALINTVDHLGSVAYKVSDLLDEKVT 96
LR QLYSAA+Y E SY N D+KQ+++E LKDYA +AL+N VDHLG+VAYK+SD+L+++
Sbjct: 1 LRPQLYSAADYCEKSYLNSDKKQMILENLKDYAARALVNAVDHLGTVAYKLSDILEKQTF 180
Query: 97 EVFGEDLRLSCIEQRIKTCQGFMDHEGHTQQSLVISTPKHHKRYILP 143
+V DL++S + QR+ TC + D EG QQ L+ P+HHK YILP
Sbjct: 181 DVSTMDLKVSTLNQRLLTCHMYTDKEGLRQQQLLAFIPRHHKHYILP 321
>AW759320
Length = 425
Score = 118 bits (296), Expect = 3e-27
Identities = 67/139 (48%), Positives = 75/139 (53%), Gaps = 50/139 (35%)
Frame = +1
Query: 91 LDEKVTEVFGEDLRLSCIEQRIKTCQGFMDHEGHTQQSLVISTPKHHKRYILP------- 143
LDEKV EV +LR+SCIEQRIKTC +MD EG TQQSLVISTPK+HKRYILP
Sbjct: 7 LDEKVVEVSVAELRVSCIEQRIKTCHEYMDREGRTQQSLVISTPKYHKRYILPGKVS*IF 186
Query: 144 -------------------------------------------VGETLHGTNSTKSKYIG 160
VGET+HG N TKSKY+G
Sbjct: 187 LYSFFLWCCFLFPFYYI*FLLLLKSMFMMLNDHIFAFLKFISSVGETMHGANRTKSKYVG 366
Query: 161 CHLDDEDDWHHFRNAVRAT 179
+L DE +W HFRNAVRAT
Sbjct: 367 YNLXDEGEWPHFRNAVRAT 423
>BE347225
Length = 450
Score = 74.3 bits (181), Expect = 7e-14
Identities = 32/99 (32%), Positives = 65/99 (65%), Gaps = 1/99 (1%)
Frame = +3
Query: 21 MHQTLLFDDSLTDLKNLRTQLYSAAEYFELSYTNDDQKQILVETLKDYAVKALINTVDHL 80
+ +T+ F+ SL +L+ LR++L+ AAEY E +++ ++K +++ K+Y + ++ VDHL
Sbjct: 114 VEETMRFEKSLQELRELRSELHKAAEYCETTFSKSEEKSDVLDNTKEYICRTMVTVVDHL 293
Query: 81 GSVAYKVSDLLDEKVTEVFGE-DLRLSCIEQRIKTCQGF 118
G+V+ + L+ T F E +LR+ C++QR+ +C+ +
Sbjct: 294 GNVSANLDGLISH--TNAFSEAELRIQCLQQRLLSCEQY 404
>BG155851 similar to GP|20197375|gb| expressed protein {Arabidopsis
thaliana}, partial (13%)
Length = 416
Score = 42.4 bits (98), Expect = 3e-04
Identities = 21/37 (56%), Positives = 26/37 (69%)
Frame = +3
Query: 13 ASNYDEVFMHQTLLFDDSLTDLKNLRTQLYSAAEYFE 49
A YDE M ++ F ++L +LKNLR QLYSAAEY E
Sbjct: 303 AMTYDEASMERSKSFVNALQELKNLRPQLYSAAEYCE 413
>TC224173 similar to UP|O81330 (O81330) F3D13.1 protein, partial (13%)
Length = 541
Score = 40.8 bits (94), Expect = 8e-04
Identities = 31/101 (30%), Positives = 50/101 (48%)
Frame = +1
Query: 181 RETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRS 240
R P++TS+ PSP+L P R SFT+ ++ ++ + S R + T + SS S
Sbjct: 151 RRRPSNTSA----PSPTLAP*RA---SFTNASLTRERWTLNSTSFCRSAPTSTATSSSSS 309
Query: 241 TTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLS 281
P + S+TP + + TSP P+ P +SA+ S
Sbjct: 310 APPTSSTSSTPTQTTCSPTSPPP-----PTSPTRSAARSAS 417
>BM143002
Length = 361
Score = 40.0 bits (92), Expect = 0.001
Identities = 18/51 (35%), Positives = 32/51 (62%)
Frame = +2
Query: 61 LVETLKDYAVKALINTVDHLGSVAYKVSDLLDEKVTEVFGEDLRLSCIEQR 111
++E K+Y +AL VDHLG+V+ ++DL+ + F + R+ C++QR
Sbjct: 155 VLENTKEYICRALFFVVDHLGNVSANINDLISQ-TNAFFEAESRIRCLKQR 304
>TC216484 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinogalactan
protein 1 precursor, partial (58%)
Length = 1213
Score = 38.1 bits (87), Expect = 0.005
Identities = 31/91 (34%), Positives = 42/91 (46%), Gaps = 2/91 (2%)
Frame = +1
Query: 173 RNAVRATIRETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSR 232
R A+ + TP+S S SPSP+ P+ SP+ RT F S
Sbjct: 109 RTTSPASSQSTPSSPPSTTTSPSPTSPPKSTA--KPPSPSAPSTTPPCRT-----FSRST 267
Query: 233 TGSMSSRSTTPKTGRSTT--PNSSNRATTSP 261
S SR+++P T STT P SS R+ T+P
Sbjct: 268 PPSTPSRTSSPSTSSSTTSAPRSSTRSPTAP 360
Score = 37.0 bits (84), Expect = 0.012
Identities = 31/91 (34%), Positives = 44/91 (48%), Gaps = 1/91 (1%)
Frame = +1
Query: 183 TPTSTSSKGNSPS-PSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRST 241
+PT T+S +S S PS P + S TSP + + +P P RT SRST
Sbjct: 100 SPTRTTSPASSQSTPSSPPSTTTSPSPTSPPKSTAKPPSPS-APSTTPPCRT---FSRST 267
Query: 242 TPKTGRSTTPNSSNRATTSPSNARVRYPSEP 272
P T T+ S++ +TTS + R P+ P
Sbjct: 268 PPSTPSRTSSPSTSSSTTSAPRSSTRSPTAP 360
>BI893904 similar to GP|15809850|gb AT5g37540/mpa22_p_70 {Arabidopsis
thaliana}, partial (29%)
Length = 421
Score = 37.7 bits (86), Expect = 0.007
Identities = 35/97 (36%), Positives = 43/97 (44%), Gaps = 17/97 (17%)
Frame = +2
Query: 183 TPTSTSSKGNSPSP------------SLQPQRVGAF----SFTSPNMAK-KDLEKRTVSP 225
TP +TSS NSPSP P+ GAF S P+ +K K RTVSP
Sbjct: 47 TPRATSSAKNSPSPLPKLPLPSYSAAPPSPETPGAFSE*TSAACPSPSKPKSPNSRTVSP 226
Query: 226 HRFPLSRTGSMSSRSTTPKTGRSTTPNSSNRATTSPS 262
P + T S RST+ T TP +S + PS
Sbjct: 227 PDNPRTTTTSQPGRSTSETT---QTPRASATSAC*PS 328
>TC232694
Length = 794
Score = 37.7 bits (86), Expect = 0.007
Identities = 31/105 (29%), Positives = 51/105 (48%), Gaps = 13/105 (12%)
Frame = -2
Query: 218 LEKRTVSPHRFPLSRTG-------SMSSRSTTPK------TGRSTTPNSSNRATTSPSNA 264
L + SPH P SR+ S+ S STTP T R++ P+S N + T+P +
Sbjct: 595 LRPQLSSPHTPPPSRSS*PNRTSLSVPSPSTTPPSPSLCYTSRTSCPSSGNSSKTTPFRS 416
Query: 265 RVRYPSEPRKSASMRLSSDVNNIRDIDQHPSKSKRLLKSLLSRRK 309
R P+ PR+ S+ N+ +P+++ +L +L+ RK
Sbjct: 415 RPPSPAAPRR*KRDEGSAPENH----PSNPARTACVLSNLVISRK 293
>TC228914 similar to UP|DAH1_MOUSE (Q9QYB2) Dachshund homolog 1 (Dach1),
partial (3%)
Length = 784
Score = 36.2 bits (82), Expect = 0.021
Identities = 35/101 (34%), Positives = 43/101 (41%), Gaps = 4/101 (3%)
Frame = +3
Query: 185 TSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPK 244
TS SS + P P+ S SP +A T S P S+ SR TTP
Sbjct: 363 TSMSSPASPPPPAPNASTSSTTSAASPALASS--AANTASATTAP--SLPSIPSR-TTPT 527
Query: 245 TGRST----TPNSSNRATTSPSNARVRYPSEPRKSASMRLS 281
T RST P SS T +P RV P+ P S++ R S
Sbjct: 528 TARSTPSSSNPTSSTSPTATPKRTRVSSPT-PSSSSTSRSS 647
Score = 29.3 bits (64), Expect = 2.5
Identities = 31/90 (34%), Positives = 43/90 (47%), Gaps = 9/90 (10%)
Frame = +3
Query: 183 TPTSTSSKGNSP--SPSLQPQRV---GAFSFTSPNMAKKDLEKRTV---SPHRFPLSRTG 234
TPTS S+ GN+P SPS +R G S S + R SP +P + G
Sbjct: 186 TPTS-SAPGNAPHSSPSASKRRPTPSGPSSAASTSPKPTSTSSRAAPSKSPFTWPSASLG 362
Query: 235 -SMSSRSTTPKTGRSTTPNSSNRATTSPSN 263
SMSS ++ P PN+S +TTS ++
Sbjct: 363 TSMSSPASPPPPA----PNASTSSTTSAAS 440
>TC232838 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;} , partial (46%)
Length = 902
Score = 35.8 bits (81), Expect = 0.027
Identities = 31/99 (31%), Positives = 48/99 (48%)
Frame = -3
Query: 184 PTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTP 243
P S+SS +S S S + S +S + + L + SP R+ + S SS S++P
Sbjct: 573 PKSSSSPCSSSSSS-------SSSSSSSESSSESLSSSSGSPPRWTSPSSSSSSSSSSSP 415
Query: 244 KTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSS 282
+G S SS+ + +SPS++ S SAS SS
Sbjct: 414 PSGAS----SSSSSLSSPSSSSSSSSSSSSSSASSTTSS 310
>TC226737 similar to UP|Q9SEE9 (Q9SEE9) Arginine/serine-rich protein, partial
(37%)
Length = 595
Score = 35.8 bits (81), Expect = 0.027
Identities = 29/98 (29%), Positives = 44/98 (44%), Gaps = 1/98 (1%)
Frame = +1
Query: 185 TSTSSKGNSPSPSLQPQRVGAFSFT-SPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTP 243
+ +SS+ +SPS S R + S + SP+ ++ P S + S SSRS +P
Sbjct: 115 SGSSSRSSSPSDSRSSSRSRSLSRSRSPSRSRSSS----------PSSSSSSRSSRSRSP 264
Query: 244 KTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLS 281
R +P + R SP R PRK + +R S
Sbjct: 265 PPQRRRSPADATRRGRSPPPQSKRASPPPRKPSPVRES 378
Score = 33.9 bits (76), Expect = 0.10
Identities = 20/56 (35%), Positives = 27/56 (47%)
Frame = +1
Query: 227 RFPLSRTGSMSSRSTTPKTGRSTTPNSSNRATTSPSNARVRYPSEPRKSASMRLSS 282
R P SSRS++P RS++ + S + SPS +R PS S S R S
Sbjct: 94 RSPSGSVSGSSSRSSSPSDSRSSSRSRSLSRSRSPSRSRSSSPSSSSSSRSSRSRS 261
>TC225167 similar to GB|CAA44226.1|20020|NTRPL121A ribosomal protein L12-1a
{Nicotiana tabacum;} , partial (70%)
Length = 993
Score = 35.4 bits (80), Expect = 0.035
Identities = 28/102 (27%), Positives = 43/102 (41%), Gaps = 1/102 (0%)
Frame = +3
Query: 179 TIRETPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSS 238
T+R +P + + SP+ P R S +SP + + P S T S S
Sbjct: 93 TLRTSPPTPPPSNSPTSPTAPPPRAAPASSSSPPSRHR---------RKSPSSATKSRDS 245
Query: 239 RSTTPKT-GRSTTPNSSNRATTSPSNARVRYPSEPRKSASMR 279
RS P+T S+ +S++ SP R PS P ++ R
Sbjct: 246 RSRKPRTSSTSSRTSSASPRQRSPP*PSPRVPSAPWRTLRRR 371
>TC206202 UP|Q99367 (Q99367) DNA-directed RNA polymerase (Fragment) ,
complete
Length = 1826
Score = 35.4 bits (80), Expect = 0.035
Identities = 30/77 (38%), Positives = 37/77 (47%), Gaps = 1/77 (1%)
Frame = +3
Query: 187 TSSKGNSP-SPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPKT 245
TSS G SP SP P G +S TSP + SP P S T S SS +P T
Sbjct: 615 TSSPGYSPSSPGYSPSSPG-YSPTSPGYSPTSPGYSPTSPGYSPTSPTYSPSSPGYSP-T 788
Query: 246 GRSTTPNSSNRATTSPS 262
+ +P S + + TSPS
Sbjct: 789 SPAYSPTSPSYSPTSPS 839
Score = 32.7 bits (73), Expect = 0.23
Identities = 29/84 (34%), Positives = 40/84 (47%), Gaps = 7/84 (8%)
Frame = +3
Query: 186 STSSKGNSP-SPSLQPQRVG------AFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSS 238
S +S G SP SP+ P G A+S TSP+ + SP P S + S +S
Sbjct: 717 SPTSPGYSPTSPTYSPSSPGYSPTSPAYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTS 896
Query: 239 RSTTPKTGRSTTPNSSNRATTSPS 262
S +P T + +P S + TSPS
Sbjct: 897 PSYSP-TSPAYSPTSPAYSPTSPS 965
Score = 31.6 bits (70), Expect = 0.51
Identities = 29/78 (37%), Positives = 37/78 (47%), Gaps = 1/78 (1%)
Frame = +3
Query: 186 STSSKGNSP-SPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPK 244
S SS G SP SP P G +S TSP + SP P S S +S + +P
Sbjct: 633 SPSSPGYSPSSPGYSPTSPG-YSPTSPGYSPTSPGYSPTSPTYSPSSPGYSPTSPAYSP- 806
Query: 245 TGRSTTPNSSNRATTSPS 262
T S +P S + + TSPS
Sbjct: 807 TSPSYSPTSPSYSPTSPS 860
>TC225340 similar to UP|Q9SPI1 (Q9SPI1) Splicing factor SR1, partial (81%)
Length = 1349
Score = 35.0 bits (79), Expect = 0.046
Identities = 43/160 (26%), Positives = 66/160 (40%), Gaps = 6/160 (3%)
Frame = +1
Query: 151 TNSTKSKYIGCHLDDEDDWHHFRNAVRATIRETPTSTSSKGNSPSPSLQPQRVGAFSFTS 210
TN KY LDD + FRNA S + +S SPS P S+ S
Sbjct: 607 TNYDDMKYAIKKLDDSE----FRNAFSKGYVRVREYDSRRDSSRSPSHGPSHSRGRSY-S 771
Query: 211 PNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTTPKTGRSTTPNSSNRATTSPSNARVRYPS 270
+ ++ R S + P ++ S S + +PK G+S+ + + + SPS +R R S
Sbjct: 772 RSRSRSHSYSRDRSQSKSPKGKS-SQRSPAKSPK-GKSSQRSPAKSPSRSPSRSRSRSRS 945
Query: 271 EPRKSASMRLSSDVN------NIRDIDQHPSKSKRLLKSL 304
+ R S + R + PS+S+ KSL
Sbjct: 946 RSLSGSRSRSRSPLPPRNKSPKKRSASRSPSRSRSRSKSL 1065
>TC227116 similar to UP|WAS2_HUMAN (Q9Y6W5) Wiskott-Aldrich syndrome protein
family member 2 (WASP-family protein member 2)
(Verprolin homology domain-containing protein 2),
partial (5%)
Length = 1154
Score = 34.7 bits (78), Expect = 0.060
Identities = 26/91 (28%), Positives = 43/91 (46%)
Frame = +3
Query: 183 TPTSTSSKGNSPSPSLQPQRVGAFSFTSPNMAKKDLEKRTVSPHRFPLSRTGSMSSRSTT 242
TPTS+SS PSP P + SF+ +T+S PL + + S ST+
Sbjct: 153 TPTSSSSWTPQPSPKTTPNPPSSTSFS-----------KTLSYSSTPLPQNPAPSPPSTS 299
Query: 243 PKTGRSTTPNSSNRATTSPSNARVRYPSEPR 273
P ++P S+ A+ + +A+ + P P+
Sbjct: 300 PP---PSSPPSTPTASFAAPSAKTKLPPTPK 383
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.128 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,910,625
Number of Sequences: 63676
Number of extensions: 176279
Number of successful extensions: 1523
Number of sequences better than 10.0: 203
Number of HSP's better than 10.0 without gapping: 1422
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1494
length of query: 323
length of database: 12,639,632
effective HSP length: 97
effective length of query: 226
effective length of database: 6,463,060
effective search space: 1460651560
effective search space used: 1460651560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC149127.4


