

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149127.2 + phase: 0
(458 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
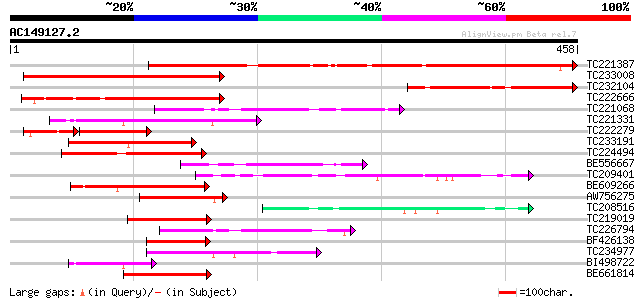
Score E
Sequences producing significant alignments: (bits) Value
TC221387 similar to UP|Q9ASW3 (Q9ASW3) AT3g49260/F2K15_120 (Guar... 371 e-103
TC233008 similar to UP|Q9ASW3 (Q9ASW3) AT3g49260/F2K15_120 (Guar... 243 1e-64
TC232104 162 2e-40
TC222666 similar to UP|Q9MAM4 (Q9MAM4) T25K16.10, partial (22%) 137 1e-32
TC221068 similar to UP|Q9LIE3 (Q9LIE3) Similarity to SF16 protei... 125 4e-29
TC221331 homologue to UP|Q9FIT1 (Q9FIT1) Emb|CAB87280.1 (AT5g620... 116 2e-26
TC222279 similar to UP|Q9ASW3 (Q9ASW3) AT3g49260/F2K15_120 (Guar... 89 3e-26
TC233191 similar to UP|Q9LK76 (Q9LK76) Arabidopsis thaliana geno... 107 9e-24
TC224494 107 9e-24
BE556667 similar to GP|11994738|db contains similarity to SF16 p... 85 6e-17
TC209401 similar to UP|Q93ZH7 (Q93ZH7) AT5g03040/F15A17_70, part... 83 3e-16
BE609266 82 4e-16
AW756275 81 1e-15
TC208516 similar to UP|Q93ZH7 (Q93ZH7) AT5g03040/F15A17_70, part... 77 2e-14
TC219019 74 1e-13
TC226794 similar to GB|AAP37767.1|30725490|BT008408 At2g43680 {A... 73 3e-13
BF426138 67 2e-11
TC234977 66 3e-11
BI498722 homologue to GP|15215590|gb| AT5g62070/mtg10_90 {Arabid... 66 3e-11
BE661814 similar to PIR|A86332|A863 F6F9.8 protein - Arabidopsis... 64 1e-10
>TC221387 similar to UP|Q9ASW3 (Q9ASW3) AT3g49260/F2K15_120 (Guard cell
associated protein) (SF16-like protein), partial (24%)
Length = 1170
Score = 371 bits (952), Expect = e-103
Identities = 204/349 (58%), Positives = 251/349 (71%), Gaps = 3/349 (0%)
Frame = +2
Query: 113 RALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQARVRARRVQLSHECLEKAMEE 172
RALRALKGLVRLQALVRGHNVRKQAQMTMRCM ALVRVQ RVRARR++L+ E L++ + E
Sbjct: 11 RALRALKGLVRLQALVRGHNVRKQAQMTMRCMHALVRVQTRVRARRLELTEEKLQRRVYE 190
Query: 173 DEEEEFVRQHETITKPMSPMRRSSVSSNNNNGWDNKCQSVKKAKENDLRKHEAAMKRERA 232
++ + V + + P+ + + +GWD++ Q+ ++ K+NDLRKHEA MKRERA
Sbjct: 191 EKVQREVDEPKQFLSPIKML--------DMDGWDSRRQTSQQIKDNDLRKHEAVMKRERA 346
Query: 233 LAYAFNYQQQQQKQHLHRNSNGDDVDMGTYHPNDDEKVQWGWNWLERWMSSQPYNVRHMV 292
LAYAFN QQQ KQH+H + NGDD+ G+Y + E+ Q WNWLERWMSSQ N+R
Sbjct: 347 LAYAFNCQQQL-KQHMHIDPNGDDI--GSYS-TERERAQLDWNWLERWMSSQSPNLR--- 505
Query: 293 PRESSYMTLPSTTSTTTDNMSEKTVEMDIMATPSRGNFNMGPMGLMAQEFHDSSPTFNRQ 352
PRE+ Y TL + TSTT D EKTVEMD+ AT + NMG + E D+SP NR
Sbjct: 506 PRETLYRTLATATSTTDDMSEEKTVEMDMGATLDSTHANMG---FINGESFDTSPISNRY 676
Query: 353 HQRPPSPGRPSYMAPTQSAKAKVRAEGPFKQRAPYGPNWNSSIKGGSIIGSGCDSSSSGG 412
HQR S G PSYMAPTQSAKAKVR++GPFKQR GP+WNSS + GS +G GCDSSSSGG
Sbjct: 677 HQRHHSAGVPSYMAPTQSAKAKVRSQGPFKQRGSPGPHWNSSTRRGSSVGLGCDSSSSGG 856
Query: 413 GTSAYKFPRSPGPKVNGVRSESRRTVGGSQD--YTEDWAVPLGA-HGWA 458
T+A+ PRSP PK+N +R +SRR GGS D + EDWA+PLGA +GWA
Sbjct: 857 PTAAHVCPRSPNPKINEIRLQSRRISGGSPDNVHIEDWALPLGATNGWA 1003
>TC233008 similar to UP|Q9ASW3 (Q9ASW3) AT3g49260/F2K15_120 (Guard cell
associated protein) (SF16-like protein), partial (29%)
Length = 700
Score = 243 bits (620), Expect = 1e-64
Identities = 134/165 (81%), Positives = 147/165 (88%), Gaps = 3/165 (1%)
Frame = +1
Query: 12 QKENKEKLEHEVAEEVSFEHFPAESSPDDVTN-EGSTTSTPV-RDDRNHAIAVAEATAAA 69
+K+N +KL+HEVAE VSFEHFPAESSPD+V+N E STTSTPV +DR+HAIAVA ATAAA
Sbjct: 196 KKDNTQKLQHEVAEVVSFEHFPAESSPDNVSNAEMSTTSTPVTNEDRSHAIAVAAATAAA 375
Query: 70 ASAAVVAAQAAARVVRLAG-YGRHNKEERAATFIQSHYRGYLARRALRALKGLVRLQALV 128
A AVVAAQAAARVVRLAG YGR +KEERAAT IQS+YRGYLARRALRALKGLVRLQALV
Sbjct: 376 AETAVVAAQAAARVVRLAGSYGRQSKEERAATLIQSYYRGYLARRALRALKGLVRLQALV 555
Query: 129 RGHNVRKQAQMTMRCMQALVRVQARVRARRVQLSHECLEKAMEED 173
RGHNVRKQAQMTMRCMQALVRVQARVRARR QLSH E+ +E+
Sbjct: 556 RGHNVRKQAQMTMRCMQALVRVQARVRARRFQLSHADQEREKKEE 690
>TC232104
Length = 540
Score = 162 bits (411), Expect = 2e-40
Identities = 86/138 (62%), Positives = 96/138 (69%), Gaps = 1/138 (0%)
Frame = +1
Query: 322 MATPSRGNF-NMGPMGLMAQEFHDSSPTFNRQHQRPPSPGRPSYMAPTQSAKAKVRAEGP 380
+ATP N N+ PM Q+F D SP NR PPSP RPSYM PTQSAKAKVRA+GP
Sbjct: 1 VATPGPTNTRNVSPMN---QDFVDLSPVSNRHRHIPPSPNRPSYMTPTQSAKAKVRAQGP 171
Query: 381 FKQRAPYGPNWNSSIKGGSIIGSGCDSSSSGGGTSAYKFPRSPGPKVNGVRSESRRTVGG 440
K R G WN S KGGS DSSSSGG AY+FPRSPGPK+NG+RS+S+R VG
Sbjct: 172 SKPRVSVG-QWNFSTKGGST----GDSSSSGGRIVAYQFPRSPGPKINGIRSQSKRIVGS 336
Query: 441 SQDYTEDWAVPLGAHGWA 458
S DYTEDW +PLGAHGWA
Sbjct: 337 SPDYTEDWTLPLGAHGWA 390
>TC222666 similar to UP|Q9MAM4 (Q9MAM4) T25K16.10, partial (22%)
Length = 1019
Score = 137 bits (344), Expect = 1e-32
Identities = 89/177 (50%), Positives = 108/177 (60%), Gaps = 13/177 (7%)
Frame = +2
Query: 10 CFQKENKEK-------------LEHEVAEEVSFEHFPAESSPDDVTNEGSTTSTPVRDDR 56
C Q+E++EK + HE S H + DV G + T D+
Sbjct: 2 CDQEEDEEKKREKRRWIFRKTHMSHEGVNNNS-NHTTQQKVQHDVAASGGGSRTD--QDQ 172
Query: 57 NHAIAVAEATAAAASAAVVAAQAAARVVRLAGYGRHNKEERAATFIQSHYRGYLARRALR 116
HA+AVA ATA AA A AQAAA V RL+ H +E AA IQ+ +RGYLARRALR
Sbjct: 173 KHALAVAMATAEAAMAT---AQAAAEVARLSKPASHAREHFAAVVIQTAFRGYLARRALR 343
Query: 117 ALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQARVRARRVQLSHECLEKAMEED 173
ALKGLV+LQALVRGHNVRKQA+MT+RCMQALVRVQARV +R++ S E K+ D
Sbjct: 344 ALKGLVKLQALVRGHNVRKQAKMTLRCMQALVRVQARVLDQRIRSSLEGSRKSTFSD 514
>TC221068 similar to UP|Q9LIE3 (Q9LIE3) Similarity to SF16 protein, partial
(33%)
Length = 652
Score = 125 bits (314), Expect = 4e-29
Identities = 80/202 (39%), Positives = 110/202 (53%)
Frame = +2
Query: 118 LKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQARVRARRVQLSHECLEKAMEEDEEEE 177
LKG+VRLQALVRGH VRKQA +T+RCMQALVRVQARVRAR H C+ A+E ++
Sbjct: 2 LKGVVRLQALVRGHAVRKQAAITLRCMQALVRVQARVRAR-----HVCM--ALETQASQQ 160
Query: 178 FVRQHETITKPMSPMRRSSVSSNNNNGWDNKCQSVKKAKENDLRKHEAAMKRERALAYAF 237
K + + GW + SV++ + L++ EAA KRERA+AYA
Sbjct: 161 ---------KHQQNLANEARVRETEEGWCDSVGSVEEIQAKILKRQEAAAKRERAMAYAL 313
Query: 238 NYQQQQQKQHLHRNSNGDDVDMGTYHPNDDEKVQWGWNWLERWMSSQPYNVRHMVPRESS 297
++Q Q S V G + P +K WGWNWLERWM+ +P+ R +
Sbjct: 314 SHQWQ-------AGSRQQPVSSGGFEP---DKNSWGWNWLERWMAVRPWENRFVDINMKD 463
Query: 298 YMTLPSTTSTTTDNMSEKTVEM 319
+T+ T D+ +E T ++
Sbjct: 464 GVTVHE--DGTKDDKNETTPQL 523
>TC221331 homologue to UP|Q9FIT1 (Q9FIT1) Emb|CAB87280.1
(AT5g62070/mtg10_90), partial (24%)
Length = 752
Score = 116 bits (290), Expect = 2e-26
Identities = 88/190 (46%), Positives = 103/190 (53%), Gaps = 19/190 (10%)
Frame = +1
Query: 33 PAESSPDDVTNEGSTTSTPVRDDRNHAIAVAEATAAAASAAVVAAQAAARVVRLAGYG-- 90
P P VT S STP+ D HAIAVA ATAA A AA+ AA AAA VVRL G
Sbjct: 187 PPPPPPSAVTYFDS--STPL-DANKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSGGVS 357
Query: 91 -----------RHNKEERAATFIQSHYRGYLARRALRALKGLVRLQALVRGHNVRKQAQM 139
R E AA IQS +RGYLARRALRALK LV+LQALVRGH VRKQ+
Sbjct: 358 ATSTRPAAMAARVGNLETAAVRIQSAFRGYLARRALRALKALVKLQALVRGHIVRKQSAD 537
Query: 140 TMRCMQALVRVQARVRARRVQLS------HECLEKAMEEDEEEEFVRQHETITKPMSPMR 193
+R MQ LVR+QA+ RA R LS + L +E E R T ++
Sbjct: 538 MLRRMQTLVRLQAQARASRAHLSDPSFNFNSSLSHYPVPEEYEHPPRGFSTKFDGSFILK 717
Query: 194 RSSVSSNNNN 203
R S ++N+ N
Sbjct: 718 RCSSNANSRN 747
>TC222279 similar to UP|Q9ASW3 (Q9ASW3) AT3g49260/F2K15_120 (Guard cell
associated protein) (SF16-like protein), partial (16%)
Length = 636
Score = 89.0 bits (219), Expect(2) = 3e-26
Identities = 47/58 (81%), Positives = 50/58 (86%)
Frame = +3
Query: 57 NHAIAVAEATAAAASAAVVAAQAAARVVRLAGYGRHNKEERAATFIQSHYRGYLARRA 114
N A+A A ATAAAA AAV AAQAAARVVRLAGYGR +KEERAA IQS+YRGYLARRA
Sbjct: 462 NPAVAFAAATAAAAEAAVAAAQAAARVVRLAGYGRQSKEERAAILIQSYYRGYLARRA 635
Score = 47.8 bits (112), Expect(2) = 3e-26
Identities = 28/47 (59%), Positives = 36/47 (76%), Gaps = 3/47 (6%)
Frame = +2
Query: 12 QKEN--KEKLEHEVAEEVSFEHFPAESSPDDVTNEGS-TTSTPVRDD 55
+KEN +E+ +H EVS EHFPAESSP D+TNEGS T+STPV ++
Sbjct: 320 KKENNKEEQWQHVAP*EVSVEHFPAESSP-DLTNEGSVTSSTPVTEE 457
>TC233191 similar to UP|Q9LK76 (Q9LK76) Arabidopsis thaliana genomic DNA,
chromosome 3, P1 clone: MDC8, partial (23%)
Length = 411
Score = 107 bits (268), Expect = 9e-24
Identities = 63/108 (58%), Positives = 77/108 (70%), Gaps = 4/108 (3%)
Frame = +3
Query: 48 TSTPVRDD-RNHAIAVAEATAAAASAAVVAAQAAARVVRLAGYGRHNK---EERAATFIQ 103
T T ++++ R HA+AVA ATA AA AAV AAQA A V+RL EE AA IQ
Sbjct: 87 TDTDIQNEQRKHAMAVAAATAVAADAAVAAAQAVAAVIRLTSTSNATSKSIEEAAAIKIQ 266
Query: 104 SHYRGYLARRALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQ 151
S +R +LA++AL AL+GLV+LQALVRGH VRKQA+ T+RCMQALV Q
Sbjct: 267 SAFRSHLAKKALCALRGLVKLQALVRGHLVRKQAKATLRCMQALVTAQ 410
>TC224494
Length = 568
Score = 107 bits (268), Expect = 9e-24
Identities = 67/119 (56%), Positives = 81/119 (67%), Gaps = 2/119 (1%)
Frame = +2
Query: 43 NEGSTTSTPVRDDRN-HAIAVAEATAAAASAAVVAA-QAAARVVRLAGYGRHNKEERAAT 100
N + S+ V D + H +A A AT AA +AAV +A + RL K+E AA
Sbjct: 230 NHSAVASSEVGSDSSPHVVAAAAATGAAFTAAVATVVRAPPKDFRLV------KQEWAAI 391
Query: 101 FIQSHYRGYLARRALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQARVRARRV 159
IQ+ +R LARRALRALKG+VR+QALVRG VRKQA +T+RCMQALVRVQARVRARRV
Sbjct: 392 RIQTAFRALLARRALRALKGVVRIQALVRGRQVRKQAAVTLRCMQALVRVQARVRARRV 568
>BE556667 similar to GP|11994738|db contains similarity to SF16
protein~gene_id:MKA23.10 {Arabidopsis thaliana}, partial
(27%)
Length = 382
Score = 85.1 bits (209), Expect = 6e-17
Identities = 52/151 (34%), Positives = 78/151 (51%)
Frame = +2
Query: 139 MTMRCMQALVRVQARVRARRVQLSHECLEKAMEEDEEEEFVRQHETITKPMSPMRRSSVS 198
+T+RCMQ+LVRVQARVRAR V++ A+E + ++Q +
Sbjct: 2 ITLRCMQSLVRVQARVRARHVRI-------ALETQATQHKLKQ---------KLANKVQV 133
Query: 199 SNNNNGWDNKCQSVKKAKENDLRKHEAAMKRERALAYAFNYQQQQQKQHLHRNSNGDDVD 258
GW + S+++ + L++ EAA KR RA+AYA ++Q Q +H +S
Sbjct: 134 RETEEGWCDSIGSIEEIQAKILKRQEAAAKRGRAMAYALSHQWQAGSRHQPVSSG----- 298
Query: 259 MGTYHPNDDEKVQWGWNWLERWMSSQPYNVR 289
+ P K WGWNWLERWM+ +P+ R
Sbjct: 299 ---FEP---YKSNWGWNWLERWMAVRPWENR 373
>TC209401 similar to UP|Q93ZH7 (Q93ZH7) AT5g03040/F15A17_70, partial (42%)
Length = 1033
Score = 82.8 bits (203), Expect = 3e-16
Identities = 79/320 (24%), Positives = 135/320 (41%), Gaps = 47/320 (14%)
Frame = +2
Query: 151 QARVRARRVQLSHECLEKAMEEDEEEEFVRQHETITKPMSPMRRSSVSSNNNNGWDNKCQ 210
Q ++R+RR+++ E ++ +++ +++H K + +R WD+ Q
Sbjct: 2 QTQIRSRRLRMLEE------NQELQKQLLQKH---AKELESIRLGEE-------WDDSIQ 133
Query: 211 SVKKAKENDLRKHEAAMKRERALAYAFNYQQQQQKQHLHRNSNGDDVDMGTYHPNDDEKV 270
S ++ + L K+EAAM+RERA+AY+F++QQ + + + + + + M +P
Sbjct: 134 SKEQVEAKLLSKYEAAMRRERAMAYSFSHQQNWK----NASRSVNPMFMDPTNP------ 283
Query: 271 QWGWNWLERWMSSQPYNVRHMVPRE--SSYMTLPSTTSTTTDNMSEKTVEMDIMATPSRG 328
WGW+WLERWM+++P+ ++ +E + S+ T+ +S+ + + +
Sbjct: 284 AWGWSWLERWMAARPWESHSLMEKEKNDNKSLRSSSRGITSAEISKSFAKFQLNSEKHSP 463
Query: 329 NFNMGPMGLMAQEFHD-----SSPTFNR-----------------------QHQRP---- 356
+ P G E H SSP R Q +RP
Sbjct: 464 TASQNP-GSPNFESHSNPPKPSSPAVARKLKKASPKDILAIDDGTKSMVSVQSERPRRHS 640
Query: 357 -------------PSPGRPSYMAPTQSAKAKVRAEGPFKQRAPYGPNWNSSIKGGSIIGS 403
SP PSYM PT+SAKAK R + PF + + G+
Sbjct: 641 IAGSIVGDDESLASSPSIPSYMVPTKSAKAKSRMQSPF------------AAENGT---- 772
Query: 404 GCDSSSSGGGTSAYKFPRSP 423
D SSG FP SP
Sbjct: 773 -PDKGSSGTAKKRLSFPASP 829
>BE609266
Length = 446
Score = 82.4 bits (202), Expect = 4e-16
Identities = 51/115 (44%), Positives = 70/115 (60%), Gaps = 3/115 (2%)
Frame = +2
Query: 50 TPVRDDRNHAIAVAEATAAAASAAVVAAQAAARVVR---LAGYGRHNKEERAATFIQSHY 106
T V + NH V ATA A V+A Q AA V+ + + EE AA IQ +
Sbjct: 101 THVESEINHD-RVEVATAVDAEEPVLAVQTAAAEVQATTIVQFDNKPTEEMAAIRIQKAF 277
Query: 107 RGYLARRALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQARVRARRVQL 161
RGYLARRALRAL+GLVRL++L+ G V++QA T+R MQ +Q ++R+RR+ +
Sbjct: 278 RGYLARRALRALRGLVRLRSLMEGPVVKRQAISTLRSMQTFAHLQTQIRSRRLTM 442
>AW756275
Length = 271
Score = 80.9 bits (198), Expect = 1e-15
Identities = 43/73 (58%), Positives = 56/73 (75%), Gaps = 2/73 (2%)
Frame = +2
Query: 106 YRGYLARRALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQARVRARRVQLSHE- 164
+R +LARRALRAL+ +VRLQA+ RG VRKQA +T+RCMQALVRVQARV AR V+ S E
Sbjct: 2 FRAFLARRALRALRAVVRLQAIFRGRLVRKQAAVTLRCMQALVRVQARVTARNVRNSPEG 181
Query: 165 -CLEKAMEEDEEE 176
++K ++E +
Sbjct: 182 KAVQKLLDEHRNQ 220
>TC208516 similar to UP|Q93ZH7 (Q93ZH7) AT5g03040/F15A17_70, partial (32%)
Length = 1059
Score = 77.0 bits (188), Expect = 2e-14
Identities = 70/264 (26%), Positives = 100/264 (37%), Gaps = 45/264 (17%)
Frame = +1
Query: 205 WDNKCQSVKKAKENDLRKHEAAMKRERALAYAFNYQQQQQKQHLHRNSNGDDVDMGTYHP 264
WD+ QS ++ + L K+EA +RERALAYAF +QQ +NS+ M
Sbjct: 10 WDDSLQSKEQIEAKLLSKYEATTRRERALAYAFTHQQN------WKNSSRSVNPMFM--- 162
Query: 265 NDDEKVQWGWNWLERWMSSQPYNVR-HMVPRESSYMTLPSTTSTTTDNMSEKTV------ 317
D WGW+WLERWM+++P+ R HM + + ++ S++ + T K+
Sbjct: 163 -DPTNPSWGWSWLERWMAARPWESRSHMDKELNDHSSIRSSSRSITGGEISKSFARFQLN 339
Query: 318 -------------EMDIMATPS-----------------RGNFNMGPMGLMAQEFHD--- 344
+TPS RG++ M H
Sbjct: 340 LEKHSPTACQNPGSPSFQSTPSKPASSSAKKPKKVSPSPRGSWVMDEDSKSLVSVHSDRF 519
Query: 345 -----SSPTFNRQHQRPPSPGRPSYMAPTQSAKAKVRAEGPFKQRAPYGPNWNSSIKGGS 399
+ + SP PSYM PTQSAKAK R + P N+ + GS
Sbjct: 520 RRHSIAGSSVRDDESLASSPAVPSYMVPTQSAKAKSRTQSPLASE-------NAKAEKGS 678
Query: 400 IIGSGCDSSSSGGGTSAYKFPRSP 423
G FP SP
Sbjct: 679 F----------GSAKKRLSFPASP 720
>TC219019
Length = 796
Score = 73.9 bits (180), Expect = 1e-13
Identities = 39/68 (57%), Positives = 50/68 (73%)
Frame = +2
Query: 96 ERAATFIQSHYRGYLARRALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQARVR 155
E AAT Q+ +RGYLARRA RALKG++RLQAL+RGH VR+QA T+ M +V+ QA VR
Sbjct: 416 EEAATKAQAAFRGYLARRAFRALKGIIRLQALIRGHLVRRQAVATLCSMYGIVKFQALVR 595
Query: 156 ARRVQLSH 163
V+ S+
Sbjct: 596 GGIVRHSN 619
>TC226794 similar to GB|AAP37767.1|30725490|BT008408 At2g43680 {Arabidopsis
thaliana;} , partial (15%)
Length = 700
Score = 72.8 bits (177), Expect = 3e-13
Identities = 48/161 (29%), Positives = 79/161 (48%), Gaps = 3/161 (1%)
Frame = +1
Query: 122 VRLQALVRGHNVRKQAQMTMRCMQALVRVQARVRARRVQLSHECLEKAMEEDEEEEFVRQ 181
VRLQ +V+G NV++Q M+ MQ LVRVQ ++++RR+Q+ + + D + +
Sbjct: 1 VRLQGVVKGQNVKRQTVNAMKHMQLLVRVQCQIQSRRIQMLEN--QARYQADFKND---- 162
Query: 182 HETITKPMSPMRRSSVSSNNNNGWDNKCQSVKKAKENDLRKHEAAMKRERALAYAFNYQQ 241
K + + S N WD+ + ++ + RK EA +KRERA+A+A+++Q
Sbjct: 163 -----KDAASILGKLTSEAGNEEWDDSLLTKEEVEARLQRKVEAIIKRERAMAFAYSHQL 327
Query: 242 QQQKQHLHRNSNGDDVDMGTYHPNDDEK---VQWGWNWLER 279
+ T+ P D + W WNWLER
Sbjct: 328 WKA------------TPKSTHTPVTDTRSGGFPWWWNWLER 414
>BF426138
Length = 438
Score = 66.6 bits (161), Expect = 2e-11
Identities = 36/52 (69%), Positives = 41/52 (78%)
Frame = +2
Query: 111 ARRALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQARVRARRVQLS 162
ARRALRALK LV+LQALVRGH VRKQ+ +R MQ LVR+QA+ RA R LS
Sbjct: 74 ARRALRALKALVKLQALVRGHIVRKQSADMLRRMQTLVRLQAQARASRAHLS 229
>TC234977
Length = 808
Score = 66.2 bits (160), Expect = 3e-11
Identities = 49/151 (32%), Positives = 78/151 (51%), Gaps = 9/151 (5%)
Frame = +2
Query: 111 ARRALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQARVRARRVQLSH-----EC 165
AR+ALRALKGLV+LQALVRGH RK+ ++ +QAL+RVQA++RA R Q+ H
Sbjct: 176 ARKALRALKGLVKLQALVRGHIERKRTAEWLQRVQALLRVQAQIRAGRAQILHSPSSTSH 355
Query: 166 LEKAMEEDEEEEFVR----QHETITKPMSPMRRSSVSSNNNNGWDNKCQSVKKAKENDLR 221
L D+ E +R +++ + P+ S N G +C+S + +D R
Sbjct: 356 LRGPATPDKFEIPIRSESMKYDQYSSPLLKRNSSKSRVQINGGNQERCRS----RSSDSR 523
Query: 222 KHEAAMKRERALAYAFNYQQQQQKQHLHRNS 252
E + R+ + +++ + L +S
Sbjct: 524 IDEQPWTQRRSWTRGCSMDEERSVRILEIDS 616
>BI498722 homologue to GP|15215590|gb| AT5g62070/mtg10_90 {Arabidopsis
thaliana}, partial (14%)
Length = 421
Score = 66.2 bits (160), Expect = 3e-11
Identities = 46/86 (53%), Positives = 50/86 (57%), Gaps = 15/86 (17%)
Frame = +1
Query: 48 TSTPVRDDRNHAIAVAEATAAAASAAVVAAQAAARVVRLAGYG---------------RH 92
+STP+ D HAIAVA ATAA A AA+ AA AAA VVRL G R
Sbjct: 166 SSTPL-DANKHAIAVAAATAAVAEAALAAAHAAAEVVRLTSGGVAATSTSSRTAVTAARV 342
Query: 93 NKEERAATFIQSHYRGYLARRALRAL 118
E AA IQS +RGYLARRALRAL
Sbjct: 343 GNLETAAVRIQSAFRGYLARRALRAL 420
>BE661814 similar to PIR|A86332|A863 F6F9.8 protein - Arabidopsis thaliana,
partial (9%)
Length = 626
Score = 63.9 bits (154), Expect = 1e-10
Identities = 35/71 (49%), Positives = 46/71 (64%)
Frame = +1
Query: 93 NKEERAATFIQSHYRGYLARRALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQA 152
N E IQ+ RG LA+R L LK +V+LQA VRGH VR+ A T+R +QA++++Q
Sbjct: 412 NPPESDVIIIQAAIRGLLAQRELLQLKKVVKLQAAVRGHLVRRHAVGTLRXIQAIIKMQI 591
Query: 153 RVRARRVQLSH 163
VRARR SH
Sbjct: 592 LVRARRALASH 624
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.127 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,876,069
Number of Sequences: 63676
Number of extensions: 277569
Number of successful extensions: 2822
Number of sequences better than 10.0: 116
Number of HSP's better than 10.0 without gapping: 2612
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2737
length of query: 458
length of database: 12,639,632
effective HSP length: 100
effective length of query: 358
effective length of database: 6,272,032
effective search space: 2245387456
effective search space used: 2245387456
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC149127.2


