

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149080.12 - phase: 0
(400 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
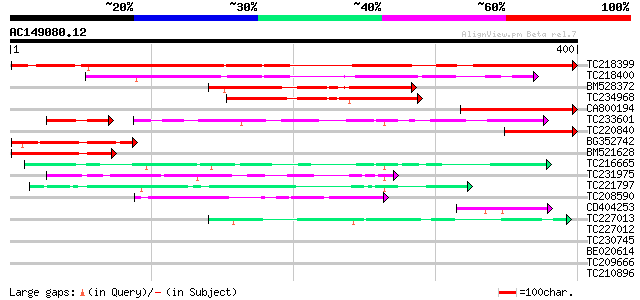
Score E
Sequences producing significant alignments: (bits) Value
TC218399 UP|PA2X_HUMAN (O15496) Group X secretory phospholipase ... 412 e-115
TC218400 similar to UP|Q8CH21 (Q8CH21) Basic protein CKT1R2, par... 213 9e-56
BM528372 170 1e-42
TC234968 similar to UP|Q9IPQ8 (Q9IPQ8) EBNA-1, partial (5%) 166 2e-41
CA800194 150 8e-37
TC233601 similar to UP|Q7XHJ1 (Q7XHJ1) Pheromone receptor-like p... 87 8e-25
TC220840 106 2e-23
BG352742 106 2e-23
BM521628 90 2e-18
TC216665 similar to UP|Q7XHJ1 (Q7XHJ1) Pheromone receptor-like p... 83 2e-16
TC231975 similar to PIR|T00986|T00986 yeast pheromone receptor-l... 77 1e-14
TC221797 homologue to PIR|T00986|T00986 yeast pheromone receptor... 57 2e-08
TC208590 similar to UP|Q6NM33 (Q6NM33) At5g19340, partial (57%) 49 3e-06
CD404253 47 2e-05
TC227013 similar to UP|GAC1_YEAST (P28006) Protein phosphatase 1... 42 4e-04
TC227012 39 0.005
TC230745 36 0.027
BE020614 36 0.035
TC209666 GB|AAB09252.1|1199563|SOYP34A 34 kDa maturing seed vacu... 35 0.060
TC210896 similar to UP|Q39540 (Q39540) AOBP (Ascorbate oxidase p... 34 0.10
>TC218399 UP|PA2X_HUMAN (O15496) Group X secretory phospholipase A2 precursor
(Phosphatidylcholine 2-acylhydrolase GX) (GX sPLA2)
(sPLA2-X) , partial (7%)
Length = 1360
Score = 412 bits (1059), Expect = e-115
Identities = 242/404 (59%), Positives = 269/404 (65%), Gaps = 5/404 (1%)
Frame = +2
Query: 2 ELQNSTKNPNLEPSNSNNGGETFFEDIDSNCSTPYVSAPSSPGRGGGPPPISS---GYFY 58
ELQ + N E N FEDIDS CSTPYVSAPSSPGRG PIS+ G+FY
Sbjct: 59 ELQTPSYNGQTE-----NPDTLLFEDIDSTCSTPYVSAPSSPGRG----PISAAGGGFFY 211
Query: 59 SAPASPLHFSITASSSYHHQTTTSSVPMS-YEFEFSARFGSTGSAASGSMTSADELFLNG 117
SAPASP+HF+ITA+S+Y H ++SS S +FEFSARF S+G SGSM+SADELFLNG
Sbjct: 212 SAPASPMHFTITAASTYTHSASSSSEKNSGCDFEFSARFVSSGFTGSGSMSSADELFLNG 391
Query: 118 QIRPMKLSSHLERPQVLAPLLDLEEEEEDEEEGEVVVVRGRDLRLRDKSVRRRTRSMSPL 177
QIRPMKLS+HLERPQVLAPLLDLEEE E+ EE E VRGRDLRLRDKS+RRRTRS+SPL
Sbjct: 392 QIRPMKLSTHLERPQVLAPLLDLEEEGEENEEVE-ASVRGRDLRLRDKSLRRRTRSLSPL 568
Query: 178 RNNSHLEWTENEDDNNNKNNVACEIEDEMNNKNNENGNDCSEMENVKVDEMGLERIEITP 237
R N+ LEWTENE+ K NV
Sbjct: 569 R-NTPLEWTENEEKTEKKENV--------------------------------------- 628
Query: 238 LDSASSSRSSSAGRSSKRWVFLKDFLRSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQ 297
S SS +GRSSKRWVFL+DFLRSKSEGRSNNKFWSTISFSP
Sbjct: 629 ----DPSVSSGSGRSSKRWVFLRDFLRSKSEGRSNNKFWSTISFSPA------------- 757
Query: 298 AQISKEKTCETQRNGSSSSSSSSSKGSWGKKMTGKPMNGVGKRRVP-PSPHELHYKANRA 356
+K+K + Q NG KK+ GKP NG+GK+RVP SPHELHYKANRA
Sbjct: 758 ---AKDKKPQNQGNGPQKP----------KKVAGKPTNGIGKKRVPAASPHELHYKANRA 898
Query: 357 QAEELRRKTFLPYRQGLLGCLGFSSKGYGAMNGFARALNPVSSR 400
QAEELRRKTFLPYRQGLLGCLGFSSKGYGAM+GFARALNPVSSR
Sbjct: 899 QAEELRRKTFLPYRQGLLGCLGFSSKGYGAMSGFARALNPVSSR 1030
>TC218400 similar to UP|Q8CH21 (Q8CH21) Basic protein CKT1R2, partial (12%)
Length = 741
Score = 213 bits (543), Expect = 9e-56
Identities = 146/322 (45%), Positives = 177/322 (54%), Gaps = 2/322 (0%)
Frame = +2
Query: 54 SGYFYSAPASPLHFSITASSSYHHQTTTSSVPMSY--EFEFSARFGSTGSAASGSMTSAD 111
+G+FYSAPASP+HF+I+A+S+Y H + SS+ + +FEFSARFGS+G SGSM+SAD
Sbjct: 5 AGFFYSAPASPMHFTISAASTYSHSPSWSSLEKNSCCDFEFSARFGSSGLTGSGSMSSAD 184
Query: 112 ELFLNGQIRPMKLSSHLERPQVLAPLLDLEEEEEDEEEGEVVVVRGRDLRLRDKSVRRRT 171
ELFLNGQIRPMKLS+HLERPQVLAPLLDLEEEEE+EE VRGRDLRLRDKS+RRRT
Sbjct: 185 ELFLNGQIRPMKLSTHLERPQVLAPLLDLEEEEEEEEN----EVRGRDLRLRDKSLRRRT 352
Query: 172 RSMSPLRNNSHLEWTENEDDNNNKNNVACEIEDEMNNKNNENGNDCSEMENVKVDEMGLE 231
RS+SPLR N+ LEWTENE + K NV
Sbjct: 353 RSLSPLR-NTPLEWTENEKKTDKKENV--------------------------------- 430
Query: 232 RIEITPLDSASSSRSSSAGRSSKRWVFLKDFLRSKSEGRSNNKFWSTISFSPTKDKKSSS 291
TP S SS +GRSSKRWVFL F + + G+ + P + K
Sbjct: 431 ----TP------SVSSGSGRSSKRWVFLDGFSKKQKRGKE**QVLVHYFLFPRRQGK--E 574
Query: 292 NNQNLQAQISKEKTCETQRNGSSSSSSSSSKGSWGKKMTGKPMNGVGKRRVPPSPHELHY 351
+AQ + K +R + + GV RV H
Sbjct: 575 GKWAAEAQEGRRKAH*RRREEARA--------------------GVVAARVAL*SH---- 682
Query: 352 KANRAQAEELRRKTFLPYRQGL 373
RAQAEELRRKTF + QGL
Sbjct: 683 ---RAQAEELRRKTFFAF*QGL 739
>BM528372
Length = 411
Score = 170 bits (430), Expect = 1e-42
Identities = 101/150 (67%), Positives = 109/150 (72%), Gaps = 3/150 (2%)
Frame = +3
Query: 141 EEEEEDEEEG---EVVVVRGRDLRLRDKSVRRRTRSMSPLRNNSHLEWTENEDDNNNKNN 197
EE+EE EEG V VVRGRDLRLRDKSVRRRTRSMSPLR+N+ LEW ENE+D+
Sbjct: 9 EEDEEVVEEGVNVNVSVVRGRDLRLRDKSVRRRTRSMSPLRSNTPLEWAENEEDH----- 173
Query: 198 VACEIEDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPLDSASSSRSSSAGRSSKRWV 257
+ + N N SE E V+ D GLE TP SASSSRSSSAGRSSKRWV
Sbjct: 174 -----DQNVTKPNKTNMISSSEAEKVE-DGFGLET---TP--SASSSRSSSAGRSSKRWV 320
Query: 258 FLKDFLRSKSEGRSNNKFWSTISFSPTKDK 287
FLKDFLRSKSEGRSNNKFWSTISFSPTKDK
Sbjct: 321 FLKDFLRSKSEGRSNNKFWSTISFSPTKDK 410
>TC234968 similar to UP|Q9IPQ8 (Q9IPQ8) EBNA-1, partial (5%)
Length = 412
Score = 166 bits (419), Expect = 2e-41
Identities = 94/140 (67%), Positives = 107/140 (76%), Gaps = 2/140 (1%)
Frame = +1
Query: 154 VVRGRDLRLRDKSVRRRTRSMSPLRNNSHLEWTENEDDNNNKNNVACEIEDEMNNKNNEN 213
VVRGRDLRLRDKSVRRRTRSMSPLR+N+ LEW ENED+ ++ + + + N N
Sbjct: 4 VVRGRDLRLRDKSVRRRTRSMSPLRSNTPLEWAENEDEEDHVH------DKNVTKPNQTN 165
Query: 214 GNDCSEMENVKVDEMGLERIEITPL--DSASSSRSSSAGRSSKRWVFLKDFLRSKSEGRS 271
SE E V+ D +GL +E TP SASSSRSSSAGRSSKRWVFLKDFLRSKSEGRS
Sbjct: 166 IISSSEAEKVE-DGLGLG-LETTPSVSGSASSSRSSSAGRSSKRWVFLKDFLRSKSEGRS 339
Query: 272 NNKFWSTISFSPTKDKKSSS 291
NNKFWSTISFSPTKDKK+ +
Sbjct: 340 NNKFWSTISFSPTKDKKNQN 399
>CA800194
Length = 441
Score = 150 bits (380), Expect = 8e-37
Identities = 70/82 (85%), Positives = 77/82 (93%)
Frame = +1
Query: 319 SSSKGSWGKKMTGKPMNGVGKRRVPPSPHELHYKANRAQAEELRRKTFLPYRQGLLGCLG 378
SSS +W ++++GKP NGVGKRRVP SPHELHYKANRAQAEELR+KTFLPYRQGLLGCLG
Sbjct: 34 SSSSQTWARRISGKPTNGVGKRRVPASPHELHYKANRAQAEELRKKTFLPYRQGLLGCLG 213
Query: 379 FSSKGYGAMNGFARALNPVSSR 400
FSSKGYGA+NGFARALNPVSSR
Sbjct: 214 FSSKGYGAINGFARALNPVSSR 279
>TC233601 similar to UP|Q7XHJ1 (Q7XHJ1) Pheromone receptor-like protein
(Fragment), partial (45%)
Length = 1014
Score = 87.0 bits (214), Expect(2) = 8e-25
Identities = 87/297 (29%), Positives = 126/297 (42%), Gaps = 4/297 (1%)
Frame = +2
Query: 88 YEFEFSARFGSTGSAASGSMTSADELFLNGQIRPMKLSSHLERPQVLAPLLDLEEEEEDE 147
+EF+FS + SA+ELFL G+IRP+K L+P + E+
Sbjct: 212 FEFDFSGQLQRPS-------LSAEELFLGGKIRPLK---------PLSPSSNKRREKRVI 343
Query: 148 EEGEVVVVRGRDLRL--RDKSVRRRTRSMSPLRNNSHLEWTENEDDNNNKNNVACEIEDE 205
EE ++ R R+++ S +PL+ + V+C+
Sbjct: 344 EESSSLLSSPPPQRKLGRERASSFSISSATPLKGRR----------GSGIQVVSCDDTSS 493
Query: 206 MNNKNNENGNDCSEMENVKVDEMGLERIEITPLDSASSSRSSSAGRSSKRWVFLKDFL-- 263
K+N + S + ++S S S G+ ++W LKDFL
Sbjct: 494 QGEKSNSSKTSES-------------------VSNSSFLSSISFGKGYRKWR-LKDFLLF 613
Query: 264 RSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQISKEKTCETQRNGSSSSSSSSSKG 323
RS SEGR+++K P + A +SK E RN S S+ SS
Sbjct: 614 RSASEGRASDK-------DPFRK----------YAVLSKTTAHEDVRNVSFRSAESSGS- 739
Query: 324 SWGKKMTGKPMNGVGKRRVPPSPHELHYKANRAQAEELRRKTFLPYRQGLLGCLGFS 380
V +RR P S HELHY NRA + EL++KT LPY+QGLLGCLGF+
Sbjct: 740 -------------VSRRRGPVSAHELHYTVNRAASVELKKKTLLPYKQGLLGCLGFN 871
Score = 44.7 bits (104), Expect(2) = 8e-25
Identities = 22/47 (46%), Positives = 30/47 (63%)
Frame = +1
Query: 27 DIDSNCSTPYVSAPSSPGRGGGPPPISSGYFYSAPASPLHFSITASS 73
+ DSNCS+PY++APSSP R G +F+SAP SP +AS+
Sbjct: 70 NFDSNCSSPYITAPSSPQRFG-------NFFFSAPTSPTRRHSSAST 189
>TC220840
Length = 542
Score = 106 bits (264), Expect = 2e-23
Identities = 50/51 (98%), Positives = 51/51 (99%)
Frame = +1
Query: 350 HYKANRAQAEELRRKTFLPYRQGLLGCLGFSSKGYGAMNGFARALNPVSSR 400
HYKANRAQAEELR+KTFLPYRQGLLGCLGFSSKGYGAMNGFARALNPVSSR
Sbjct: 10 HYKANRAQAEELRKKTFLPYRQGLLGCLGFSSKGYGAMNGFARALNPVSSR 162
>BG352742
Length = 355
Score = 106 bits (264), Expect = 2e-23
Identities = 59/92 (64%), Positives = 66/92 (71%), Gaps = 3/92 (3%)
Frame = +1
Query: 2 ELQNST---KNPNLEPSNSNNGGETFFEDIDSNCSTPYVSAPSSPGRGGGPPPISSGYFY 58
+LQ T N N E SNSNNG + FFED DS CSTPYVSAPSSPGRG P G+FY
Sbjct: 103 QLQTPTTTHNNTNPETSNSNNG-DLFFEDFDSTCSTPYVSAPSSPGRGPLP-----GFFY 264
Query: 59 SAPASPLHFSITASSSYHHQTTTSSVPMSYEF 90
SAPASP+HF+ITA+SSY + T SS PM YEF
Sbjct: 265 SAPASPMHFAITAASSY--ENTPSSAPMGYEF 354
>BM521628
Length = 428
Score = 89.7 bits (221), Expect = 2e-18
Identities = 45/74 (60%), Positives = 52/74 (69%)
Frame = +2
Query: 2 ELQNSTKNPNLEPSNSNNGGETFFEDIDSNCSTPYVSAPSSPGRGGGPPPISSGYFYSAP 61
+LQ T N +NNG FF+D DS CSTPYVSAPSSPGRG P G+FYSAP
Sbjct: 215 QLQTPTITHNNTNPETNNGESLFFQDFDSTCSTPYVSAPSSPGRGPLP-----GFFYSAP 379
Query: 62 ASPLHFSITASSSY 75
ASP+HF+ITA+SSY
Sbjct: 380 ASPMHFAITAASSY 421
>TC216665 similar to UP|Q7XHJ1 (Q7XHJ1) Pheromone receptor-like protein
(Fragment), partial (42%)
Length = 1006
Score = 83.2 bits (204), Expect = 2e-16
Identities = 107/398 (26%), Positives = 153/398 (37%), Gaps = 26/398 (6%)
Frame = +2
Query: 11 NLEPSNSNNGGETFFEDIDSNCSTPYVSAPSSPGRGGGPPPISSGYFYSAPASPLHFSIT 70
N+E +S++ D + +PY+SAPSSP R G Y+ SAP+SP
Sbjct: 2 NMEVLSSHSPSSNDPFDFNRARISPYLSAPSSPKRFG------QFYYLSAPSSP------ 145
Query: 71 ASSSYHHQTTTSSVPMSYEFEFSAR----FGSTGSAASGSMTSADELFLNGQIRPMKLSS 126
S +F+ +A F + S S SA ELF G+I+P+
Sbjct: 146 ----------------SKDFDLAAHDDDAFAFFVTGESKSPRSAAELFDGGKIKPLSEDD 277
Query: 127 HLERPQVLAPLLDLE-------------------EEEEDEEEGEVVVVRGRDLRLRDKSV 167
E + +PLL + ++EE + E RGRD R S
Sbjct: 278 SSESAK--SPLLPAKRSSIAQGKKAIREAFSTRKKKEEGDLSEEEQETRGRD---RTASS 442
Query: 168 RRRTRSMSPLRNNSHLEWTENEDDNNNKNNVACEIEDEMNNKNNENGNDCSEMENVKVDE 227
R TRS+SP SH W DE N++ N
Sbjct: 443 SRVTRSLSPCNRISHYTW------------------DEERNQSQSN-------------- 526
Query: 228 MGLERIEITPLDSASSSRSSSAGRSSKRWVFLKDFL--RSKSEGRSNNKFWSTISFSPTK 285
++ S ++S+RW L DFL RS SEGR ++K + PT
Sbjct: 527 -----------------KADSLSKTSRRWR-LTDFLLFRSASEGRGSSK--DPLRKFPTF 646
Query: 286 DKKSSSNNQNLQAQISKEKTCETQRNGSSSSSSSSSKGSWGKKMTGKPMNGVGKRRVPPS 345
KK ++ +A KT +R+ P S
Sbjct: 647 YKKP----EDPKASNPGPKT---------------------------------RRKEPLS 715
Query: 346 PHELHYKANRAQAEELRRKTFLPYRQGLLGCL-GFSSK 382
HELHY +A+ E+L+++TFLPY+QG+LG L GF SK
Sbjct: 716 AHELHYARKKAETEDLKKRTFLPYKQGILGRLAGFGSK 829
>TC231975 similar to PIR|T00986|T00986 yeast pheromone receptor-like protein
AR781 [imported] - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (22%)
Length = 799
Score = 77.0 bits (188), Expect = 1e-14
Identities = 78/271 (28%), Positives = 117/271 (42%), Gaps = 23/271 (8%)
Frame = +1
Query: 27 DIDSNCSTPYVSAPSSPGRGGGPPPISSGYFYSAPASPLHFSITASSSYHHQTTTSSVPM 86
+ DSNCS+P+++APSSP P +F+SAP SP +SS+ H + +SS +
Sbjct: 112 NFDSNCSSPFITAPSSPQFFASNKP---NFFFSAPTSPTR----GTSSFFHDSPSSSSVL 270
Query: 87 SYEFEFSARFGSTGSAASGSMTSADELFLNGQIRPMKLSSHLERP--------------- 131
+E + F +G S+ SA ELF G+IRP+K L+ P
Sbjct: 271 PHEEQQDFEFNFSGHLDRPSL-SAAELFDGGKIRPLKPPPRLQTPVTSPRSKLSPRKKKK 447
Query: 132 ------QVLAPLLDLEEEEEDEEEGEVVVVRGRDLRLRDKSVRRRTRSMSPLRNNSHLEW 185
+ + L EE+EE + E V V G R+ +RS+SPLR +
Sbjct: 448 DFDPFAEAMKETLKREEQEETQRRRERVSVSG----------RKGSRSLSPLRIS----- 582
Query: 186 TENEDDNNNKNNVACEIEDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPLDSASSSR 245
+ V C+ ED+ + + N N S +SS
Sbjct: 583 ----------DIVVCDSEDKSVSSSTSNNN------------------------SKTSSF 660
Query: 246 SSSAGRSSKRWVFLKDFL--RSKSEGRSNNK 274
SS +++W F +DFL RS SEGR+ +K
Sbjct: 661 LSSI-PFTRKWRF-RDFLLFRSASEGRATDK 747
>TC221797 homologue to PIR|T00986|T00986 yeast pheromone receptor-like
protein AR781 [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (16%)
Length = 763
Score = 56.6 bits (135), Expect = 2e-08
Identities = 86/325 (26%), Positives = 123/325 (37%), Gaps = 13/325 (4%)
Frame = +3
Query: 15 SNSNNGGETFFEDIDSNCSTPYVSAPSSPGRGGGPPPISSGYFYSAPASPLHFS-ITASS 73
S S+ +TF D N + + S PSSP +G G Y+ SAPASP S +
Sbjct: 3 SPSSPNMDTF--DFGGNMGSIFQSVPSSP-KGFG------NYYLSAPASPSRMSELYTEF 155
Query: 74 SYHHQTTTSSVPMSYEFE----------FSARFGSTGSAASGSMTSADELFLNGQIRPMK 123
Y+ + SS + + +S F + S SA+ELF G+I+P+
Sbjct: 156 EYYSSASPSSFEAGNKIDDDDEDENNNDYSFAFYVNRDQSDKSSRSAEELFDGGKIKPLN 335
Query: 124 LSSHLERPQVLAPLLDLEEEEEDEEEGEVVVVRGRDLRLRDKSVRRRTRSMSPLRNNSHL 183
SS R +A E++ E E L + VRR RS+SP R + +
Sbjct: 336 ESS---RATSVA-----EQKRGRERERSSSSTTTTKLSSSNSGVRRVARSLSPYRKSWEV 491
Query: 184 EWTENEDDNNNKNNVACEIEDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPLDSASS 243
E + + +K + + SS
Sbjct: 492 EQQKEQPRIISKEESSASV--------------------------------------LSS 557
Query: 244 SRSSSAGRSSKRWVFLKDFL--RSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQIS 301
S SSS G SKRW LKDFL RS SEGR ++K L+ +
Sbjct: 558 STSSSKG--SKRW-SLKDFLLFRSASEGRGSSK-------------------DALKKYYN 671
Query: 302 KEKTCETQRNGSSSSSSSSSKGSWG 326
K+ + E + SS SS SS+ G
Sbjct: 672 KKNSNEEVKGSSSFRSSDSSRSRKG 746
>TC208590 similar to UP|Q6NM33 (Q6NM33) At5g19340, partial (57%)
Length = 1103
Score = 49.3 bits (116), Expect = 3e-06
Identities = 51/180 (28%), Positives = 83/180 (45%), Gaps = 1/180 (0%)
Frame = +1
Query: 89 EFEFSARFGSTGSAASGSMTSADELFLNGQIRPMKLSSHLER-PQVLAPLLDLEEEEEDE 147
EFEF S ++ + ++ +ADELF G++ P HLE+ ++ + EEEEE+E
Sbjct: 280 EFEFL----SNNTSNNNTVVTADELFFEGKLLPFWQMQHLEKLSKINLKTKEGEEEEEEE 447
Query: 148 EEGEVVVVRGRDLRLRDKSVRRRTRSMSPLRNNSHLEWTENEDDNNNKNNVACEIEDEMN 207
E EVVVV NN N++DNNN N+ + + ++
Sbjct: 448 LEEEVVVV----------------------SNN-------NKEDNNNSNS-SSRVNWFVD 537
Query: 208 NKNNENGNDCSEMENVKVDEMGLERIEITPLDSASSSRSSSAGRSSKRWVFLKDFLRSKS 267
+ + C+ + + + L++ + L +SSS SSS+ SS V K+ RS S
Sbjct: 538 DDPSPRPPKCTVLWK---ELLRLKKQRASSLSPSSSSSSSSSSASSLGDVAAKEGSRSSS 708
>CD404253
Length = 617
Score = 46.6 bits (109), Expect = 2e-05
Identities = 29/76 (38%), Positives = 43/76 (56%), Gaps = 8/76 (10%)
Frame = -3
Query: 316 SSSSSSKGSWGKKMTGKPM---NGVGKRRVPPSP----HELHYKANRAQAEELRRKTFLP 368
SS +S+K K TGK +G G++R+ P HE Y NR E++R+++LP
Sbjct: 546 SSVNSAKKKGTKSETGKKSPASSGGGEKRIVAVPAVSAHEALYVRNREMRREVKRRSYLP 367
Query: 369 YRQGLLG-CLGFSSKG 383
YRQ L+G C+ +S G
Sbjct: 366 YRQDLVGLCVNLNSMG 319
>TC227013 similar to UP|GAC1_YEAST (P28006) Protein phosphatase 1 regulatory
subunit GAC1, partial (3%)
Length = 1149
Score = 42.4 bits (98), Expect = 4e-04
Identities = 60/265 (22%), Positives = 97/265 (35%), Gaps = 9/265 (3%)
Frame = +2
Query: 141 EEEEEDEEEGEVVVVR--GRDLRLRDKSVRRRTRSMSPLRNNSHLEWTENEDDNNNKNNV 198
EEEEE+EEE V+ G + D + R + P+ N
Sbjct: 254 EEEEEEEEEFSFVLTNSDGSPISADDAFDNGQIRPIFPIFN------------------- 376
Query: 199 ACEIEDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPLDSA-------SSSRSSSAGR 251
+D + + + + G + V V+ E P + A ++ +S+S G
Sbjct: 377 ----QDLLFSDDYDGGEGLRQPIKVFVEHKDEFLSEAAPAEGAYCEWNPKAAVKSNSTG- 541
Query: 252 SSKRWVFLKDFLRSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQISKEKTCETQRN 311
SK W F LRS S+G+ F + + + +K S + K KT T
Sbjct: 542 FSKLWRFRDVKLRSNSDGKDAFVFLNHAAPAKPAEKARS-------VVVKKGKTTTTTAA 700
Query: 312 GSSSSSSSSSKGSWGKKMTGKPMNGVGKRRVPPSPHELHYKANRAQAEELRRKTFLPYRQ 371
S+ HE HY +RA+ E +RK++LPY+Q
Sbjct: 701 ASA--------------------------------HEKHYIMSRARKESDKRKSYLPYKQ 784
Query: 372 GLLGCLGFSSKGYGAMNGFARALNP 396
+LG + NG +R ++P
Sbjct: 785 NMLGF-------FANANGLSRNVHP 838
Score = 28.5 bits (62), Expect = 5.6
Identities = 16/47 (34%), Positives = 22/47 (46%)
Frame = +2
Query: 76 HHQTTTSSVPMSYEFEFSARFGSTGSAASGSMTSADELFLNGQIRPM 122
H++ E E F + + GS SAD+ F NGQIRP+
Sbjct: 221 HNEDEIEEEEEEEEEEEEEEFSFVLTNSDGSPISADDAFDNGQIRPI 361
>TC227012
Length = 931
Score = 38.5 bits (88), Expect = 0.005
Identities = 20/52 (38%), Positives = 30/52 (57%)
Frame = +3
Query: 345 SPHELHYKANRAQAEELRRKTFLPYRQGLLGCLGFSSKGYGAMNGFARALNP 396
S HE HY +RA+ E +RK++LPY+Q L G +S G +R ++P
Sbjct: 693 SAHEKHYVMSRARKESDKRKSYLPYKQDLFGFFANAS-------GLSRNVHP 827
>TC230745
Length = 932
Score = 36.2 bits (82), Expect = 0.027
Identities = 21/79 (26%), Positives = 38/79 (47%)
Frame = +2
Query: 69 ITASSSYHHQTTTSSVPMSYEFEFSARFGSTGSAASGSMTSADELFLNGQIRPMKLSSHL 128
+ + + H+ P+S +FEFS + + SM SADE+F G + P+K
Sbjct: 113 VVSQQAIKHENIYREDPVSSDFEFSVK--------NNSMISADEVFFQGMLLPLKSDCSN 268
Query: 129 ERPQVLAPLLDLEEEEEDE 147
++ + LL ++ E+E
Sbjct: 269 KKVTLRDELLVNDDHYEEE 325
>BE020614
Length = 392
Score = 35.8 bits (81), Expect = 0.035
Identities = 22/61 (36%), Positives = 33/61 (54%)
Frame = +1
Query: 89 EFEFSARFGSTGSAASGSMTSADELFLNGQIRPMKLSSHLERPQVLAPLLDLEEEEEDEE 148
EFEF S ++++ ++ +ADELF G++ P HLE+ + EEEE EE
Sbjct: 220 EFEFL----SNNTSSNNTVLTADELFFEGKLLPFWQMQHLEKLSKINLKTKEGEEEELEE 387
Query: 149 E 149
E
Sbjct: 388 E 390
>TC209666 GB|AAB09252.1|1199563|SOYP34A 34 kDa maturing seed vacuolar thiol
protease precursor {Glycine max;} , partial (8%)
Length = 572
Score = 35.0 bits (79), Expect = 0.060
Identities = 25/77 (32%), Positives = 36/77 (46%)
Frame = +2
Query: 58 YSAPASPLHFSITASSSYHHQTTTSSVPMSYEFEFSARFGSTGSAASGSMTSADELFLNG 117
Y+ P+SP H + SSS S +FEF+ S S ++ ADELF G
Sbjct: 167 YTLPSSPSHSFSSRSSS------------SSDFEFTISVSPRKS--STALCPADELFYKG 304
Query: 118 QIRPMKLSSHLERPQVL 134
Q+ P+ LS + + L
Sbjct: 305 QLLPLHLSQRISMVRTL 355
>TC210896 similar to UP|Q39540 (Q39540) AOBP (Ascorbate oxidase
promoter-binding protein), partial (15%)
Length = 633
Score = 34.3 bits (77), Expect = 0.10
Identities = 17/38 (44%), Positives = 21/38 (54%), Gaps = 4/38 (10%)
Frame = +1
Query: 192 NNNKNNVACEIEDEMN----NKNNENGNDCSEMENVKV 225
NNN N IED+ N N NN NG+DCS ++ V
Sbjct: 175 NNNTRNGFHTIEDQRNKDNHNNNNNNGDDCSSASSITV 288
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.307 0.125 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,308,430
Number of Sequences: 63676
Number of extensions: 298717
Number of successful extensions: 3841
Number of sequences better than 10.0: 157
Number of HSP's better than 10.0 without gapping: 2816
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3424
length of query: 400
length of database: 12,639,632
effective HSP length: 99
effective length of query: 301
effective length of database: 6,335,708
effective search space: 1907048108
effective search space used: 1907048108
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC149080.12


