

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149079.7 + phase: 0
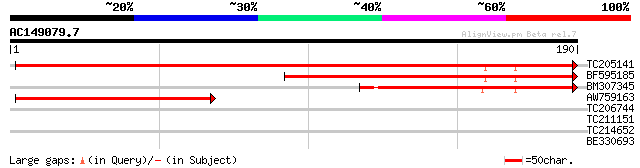
(190 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC205141 weakly similar to UP|Q86BN8 (Q86BN8) CG10371-PB, partia... 243 4e-65
BF595185 similar to GP|20197531|gb| expressed protein {Arabidops... 137 2e-33
BM307345 102 1e-22
AW759163 weakly similar to GP|14090343|dbj P0638D12.10 {Oryza sa... 74 3e-14
TC206744 similar to UP|Q6NKR2 (Q6NKR2) At5g56610, partial (75%) 38 0.003
TC211151 UP|Q8LJR8 (Q8LJR8) RING-H2 finger protein, complete 28 3.3
TC214652 homologue to UP|Q39860 (Q39860) GTPase, complete 27 4.4
BE330693 26 9.7
>TC205141 weakly similar to UP|Q86BN8 (Q86BN8) CG10371-PB, partial (15%)
Length = 1915
Score = 243 bits (620), Expect = 4e-65
Identities = 140/190 (73%), Positives = 151/190 (78%), Gaps = 2/190 (1%)
Frame = +1
Query: 3 KMHYLDALHMSTAKLDGVAAQLSLFVIWCTTN*CHLMLHMNM*SQFVQGCF*LLLSGKLS 62
KMH D LHMS AKLD V A+L LFVIWCTT * HLMLHM M*+QF QGCF*L LSGKLS
Sbjct: 979 KMHCRDELHMSIAKLDVVVARLLLFVIWCTTK**HLMLHMLM*NQFDQGCF*LPLSGKLS 1158
Query: 63 RNTTATLL*GGLLDLHPQLNYL*RLLSLQQLHKILYLLMTIL*LW*QSKISKDTTQVVNQ 122
+NTT L *GG LD+ Q YL*+LL QQ+H+IL LMTIL* W*QS+I K TTQVVNQ
Sbjct: 1159 KNTTTILW*GGPLDVLLQPIYL*KLLRWQQVHEIL*CLMTIL*SW*QSQI*KVTTQVVNQ 1338
Query: 123 IPRQEKYGQI*VLFTVCELLDRQHWLEYHAFGCDMV-PTRRYLREN*-AWKTVAQSELIT 180
P Q KYGQI*VL+TV ELLDRQHW EYHAFGCDMV PTRR+LR N* A K VAQS LIT
Sbjct: 1339 EPWQVKYGQI*VLYTVYELLDRQHWREYHAFGCDMVLPTRRFLRRN*AAGKAVAQSGLIT 1518
Query: 181 WEKLMLISMC 190
WEKL+LIS C
Sbjct: 1519 WEKLVLISTC 1548
>BF595185 similar to GP|20197531|gb| expressed protein {Arabidopsis
thaliana}, partial (13%)
Length = 418
Score = 137 bits (346), Expect = 2e-33
Identities = 77/100 (77%), Positives = 83/100 (83%), Gaps = 2/100 (2%)
Frame = +3
Query: 93 LHKILYLLMTIL*LW*QSKISKDTTQVVNQIPRQEKYGQI*VLFTVCELLDRQHWLEYHA 152
+H+IL LMTIL* W*QS+I K TTQVVNQ P Q KYGQI*VL+TV ELLDRQHW EYHA
Sbjct: 3 VHEIL*CLMTIL*SW*QSQI*KVTTQVVNQEPWQVKYGQI*VLYTVYELLDRQHWREYHA 182
Query: 153 FGCDMV-PTRRYLREN*-AWKTVAQSELITWEKLMLISMC 190
FGCDMV PTRR+LR N* A K VAQS LITWEKL+LIS C
Sbjct: 183 FGCDMVLPTRRFLRRN*AAGKAVAQSGLITWEKLVLISTC 302
>BM307345
Length = 355
Score = 102 bits (253), Expect = 1e-22
Identities = 57/75 (76%), Positives = 61/75 (81%), Gaps = 2/75 (2%)
Frame = +2
Query: 118 QVVNQIPRQEKYGQI*VLFTVCELLDRQHWLEYHAFGCDM-VPTRRYLREN*-AWKTVAQ 175
QVVNQ P Q KYGQI*VL+ V ELLDRQHW EYHAFGCDM +PTRR+ R N* A K VAQ
Sbjct: 2 QVVNQ-PWQVKYGQI*VLYIVYELLDRQHWREYHAFGCDMLLPTRRFPRRN*AAGKAVAQ 178
Query: 176 SELITWEKLMLISMC 190
S LITWEKL+LIS C
Sbjct: 179 SGLITWEKLVLISTC 223
>AW759163 weakly similar to GP|14090343|dbj P0638D12.10 {Oryza sativa
(japonica cultivar-group)}, partial (14%)
Length = 333
Score = 74.3 bits (181), Expect = 3e-14
Identities = 42/67 (62%), Positives = 51/67 (75%)
Frame = +3
Query: 3 KMHYLDALHMSTAKLDGVAAQLSLFVIWCTTN*CHLMLHMNM*SQFVQGCF*LLLSGKLS 62
K+ YL+ LHMS KL+ V +QL LF+I TT * HL+LH+ M*+QF Q CF*L LSGKLS
Sbjct: 129 KIRYLNELHMSITKLNMVISQLLLFMI*YTTK**HLILHILM*NQFDQECF*LPLSGKLS 308
Query: 63 RNTTATL 69
+NTTA L
Sbjct: 309 KNTTAIL 329
>TC206744 similar to UP|Q6NKR2 (Q6NKR2) At5g56610, partial (75%)
Length = 1164
Score = 37.7 bits (86), Expect = 0.003
Identities = 33/94 (35%), Positives = 43/94 (45%), Gaps = 3/94 (3%)
Frame = +2
Query: 3 KMHYLDALHMSTAKLDGVAAQLSLFVIWCTTN*CHLMLHMNM*SQFVQGCF*LLLSGKLS 62
+M L M T KL G Q FVIW + HL+LH NM C+* SG+L
Sbjct: 500 RMQLAAKLLMFTVKLGGGGVQQLCFVIWLNIST*HLLLHWNMFGLEDLECY*HHHSGRLF 679
Query: 63 RNTTATLL*GGLLDLHPQ---LNYL*RLLSLQQL 93
+ T+ GLL H + YL* L L+ +
Sbjct: 680 KTITSE----GLLLCHTPPLGMQYL*PKLILKAI 769
>TC211151 UP|Q8LJR8 (Q8LJR8) RING-H2 finger protein, complete
Length = 1107
Score = 27.7 bits (60), Expect = 3.3
Identities = 10/23 (43%), Positives = 14/23 (60%)
Frame = -3
Query: 139 CELLDRQHWLEYHAFGCDMVPTR 161
C LDR+H++ AF C + P R
Sbjct: 205 CRALDREHFVPIDAFWCGIAPKR 137
>TC214652 homologue to UP|Q39860 (Q39860) GTPase, complete
Length = 1004
Score = 27.3 bits (59), Expect = 4.4
Identities = 12/24 (50%), Positives = 15/24 (62%)
Frame = +1
Query: 7 LDALHMSTAKLDGVAAQLSLFVIW 30
+ A H ST KL + LSLFV+W
Sbjct: 4 ITAKHASTTKLSFPSLSLSLFVVW 75
>BE330693
Length = 247
Score = 26.2 bits (56), Expect = 9.7
Identities = 13/38 (34%), Positives = 20/38 (52%), Gaps = 3/38 (7%)
Frame = +2
Query: 9 ALHM---STAKLDGVAAQLSLFVIWCTTN*CHLMLHMN 43
ALH+ K+D A +LF+IW + C + +H N
Sbjct: 2 ALHLFDRDCYKIDPFARSKTLFLIWSHAHKCFMKMHNN 115
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.353 0.154 0.539
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,644,123
Number of Sequences: 63676
Number of extensions: 111868
Number of successful extensions: 1017
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 1010
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1011
length of query: 190
length of database: 12,639,632
effective HSP length: 92
effective length of query: 98
effective length of database: 6,781,440
effective search space: 664581120
effective search space used: 664581120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC149079.7


