

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149079.6 - phase: 0
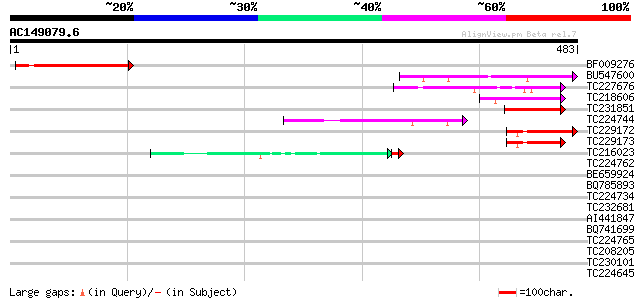
(483 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BF009276 145 3e-35
BU547600 homologue to GP|21554414|gb| glycoprotein homolog {Arab... 113 2e-25
TC227676 49 5e-06
TC218606 48 9e-06
TC231851 similar to UP|Q94F56 (Q94F56) AT5g56980/MHM17_10, parti... 46 4e-05
TC224744 similar to UP|Q09085 (Q09085) Hydroxyproline-rich glyco... 43 4e-04
TC229172 similar to UP|Q8IP68 (Q8IP68) CG31813-PA, partial (8%) 42 6e-04
TC229173 42 8e-04
TC216023 similar to GB|AAM78036.1|21928015|AY125526 At2g01100/F2... 37 9e-04
TC224762 homologue to UP|Q09085 (Q09085) Hydroxyproline-rich gly... 40 0.003
BE659924 39 0.004
BQ785893 similar to PIR|T10863|T108 extensin precursor - kidney ... 39 0.004
TC224734 homologue to UP|Q09085 (Q09085) Hydroxyproline-rich gly... 39 0.005
TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g4... 39 0.007
AI441847 homologue to PIR|T11622|T11 extensin class 1 precursor ... 39 0.007
BQ741699 homologue to GP|3204132|emb extensin {Cicer arietinum},... 38 0.009
TC224765 similar to UP|Q09082 (Q09082) Extensin (Class I), parti... 38 0.009
TC208205 hydroxyproline-rich glycoprotein 38 0.009
TC230101 similar to UP|Q41707 (Q41707) Extensin class 1 protein ... 38 0.009
TC224645 homologue to UP|Q41707 (Q41707) Extensin class 1 protei... 38 0.009
>BF009276
Length = 427
Score = 145 bits (367), Expect = 3e-35
Identities = 73/101 (72%), Positives = 86/101 (84%), Gaps = 1/101 (0%)
Frame = +2
Query: 6 SIQITKHQKPTLDNVKQNQQEPNKFHYNFAYKATIVLIFFVILPLLPSQAPEFISQNLLT 65
+I TKHQK L Q+Q++ NKF+Y+F YKAT+VLIFFVILPL PSQAPEFI+Q+LLT
Sbjct: 134 AIHATKHQKLPL---AQHQEDQNKFYYHFLYKATLVLIFFVILPLFPSQAPEFINQSLLT 304
Query: 66 RNWELLHLLFVGIAISYGLFSRRNQEPDKD-NNNNTKFDNA 105
RNWELLHLLFVG+AISYGLFSRRN E +K+ NNNN+KFD A
Sbjct: 305 RNWELLHLLFVGVAISYGLFSRRNDETEKEHNNNNSKFDTA 427
>BU547600 homologue to GP|21554414|gb| glycoprotein homolog {Arabidopsis
thaliana}, partial (6%)
Length = 659
Score = 113 bits (283), Expect = 2e-25
Identities = 75/173 (43%), Positives = 97/173 (55%), Gaps = 22/173 (12%)
Frame = -2
Query: 333 PPPPTMFYKSTFLKSRYGE----------------KNGRNMSMGKSIEEGTNEKED---- 372
PPPP MF+KS +K RYG K+ RNM +GK I+E +
Sbjct: 658 PPPPMMFHKSVSMKPRYGGSFNEESSFNKELKRSFKSERNMPVGKKIDEENKPMQRTSFR 479
Query: 373 EDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSS 432
D+ S + + +KE F+ + E E+T++ EDEDVG + V++ +
Sbjct: 478 SDKIMGHASVPLVSQPAEKESFLVESNDDEDEDTET-EDEDVGGGRIIVQNQNNDSGKGP 302
Query: 433 SLSGGID--TVSDEGPDVDKKADEFIAKFREQIRLQRIESIKRSTRVARNSSR 483
+ GG T DEGPDVDKKADEFIAKFREQIRLQRIE IKRST+ ARNS+R
Sbjct: 301 QVIGGESSKTDGDEGPDVDKKADEFIAKFREQIRLQRIECIKRSTKSARNSTR 143
>TC227676
Length = 1104
Score = 48.9 bits (115), Expect = 5e-06
Identities = 44/170 (25%), Positives = 80/170 (46%), Gaps = 24/170 (14%)
Frame = +1
Query: 328 PPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIE-EGTNEKEDEDEEEKEYSTSSMQ 386
P P P PP + + L+ ++ N+S+ K ++ E E E E E+ + + S+
Sbjct: 340 PQPQPEKPPELLRTPSLLERL---RSFHNLSLHKHVQPEPEPEPEPEPEKPELVRSPSLL 510
Query: 387 EQTDKERF--IEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGG------- 437
++ F + + ++ D D D G + S K+ + + S+S+ GG
Sbjct: 511 QRIQSINFSHLYRSDFSHRDDEDPDSGSDPG--RGSGKAAEMRK--SASVRGGLTDSEWE 678
Query: 438 ------------IDTVSD--EGPDVDKKADEFIAKFREQIRLQRIESIKR 473
++T + E +VD KAD+FI +F++Q+RLQRI+S+ R
Sbjct: 679 EVEKRRPQTARPVETTTSWREDEEVDAKADDFINRFKKQLRLQRIDSLLR 828
>TC218606
Length = 1122
Score = 48.1 bits (113), Expect = 9e-06
Identities = 30/75 (40%), Positives = 43/75 (57%), Gaps = 2/75 (2%)
Frame = +1
Query: 401 LEAEETDSDEDE--DVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAK 458
+E E+ DS + + + Q +KS + EP S GG E VD KAD+FI +
Sbjct: 631 VEMEKPDSIQQQLTRAPSILQRLKSSLSFEPESEVTGGGELAEETEEEGVDAKADDFINR 810
Query: 459 FREQIRLQRIESIKR 473
FR+Q+RLQR++SI R
Sbjct: 811 FRQQLRLQRLDSIIR 855
>TC231851 similar to UP|Q94F56 (Q94F56) AT5g56980/MHM17_10, partial (12%)
Length = 506
Score = 45.8 bits (107), Expect = 4e-05
Identities = 24/52 (46%), Positives = 32/52 (61%)
Frame = +1
Query: 422 KSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAKFREQIRLQRIESIKR 473
+S+ EP S GG E VD KAD+FI +FR+Q+RLQR++SI R
Sbjct: 136 RSMSVTEPESEVTGGGELVEETEEEGVDAKADDFINRFRQQLRLQRLDSIIR 291
>TC224744 similar to UP|Q09085 (Q09085) Hydroxyproline-rich glycoprotein
(HRGP) (Fragment), partial (30%)
Length = 592
Score = 42.7 bits (99), Expect = 4e-04
Identities = 40/166 (24%), Positives = 69/166 (41%), Gaps = 9/166 (5%)
Frame = +1
Query: 234 EDKLKQKEEENAVLPS-PIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASK 292
E K+K K++ VL + P + S+ +PK P ++
Sbjct: 100 ELKVKSKDKYEVVLKAKPFAYASKKHFEECYKPKPS-------------PTPYYYKSPPP 240
Query: 293 PSMVKTSSIKFTPSES-VAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYK-----STFLK 346
PS + PS + V K+ +K +YY+S PPP P P P +YK S K
Sbjct: 241 PSPIYIYKSPPPPSPTYVYKSPPPPVKSPPYYYQSPPPPSPKPKPPYYYKSPPPPSPKPK 420
Query: 347 SRYGEKNGRNMSMGKSIEEGTNEKE--DEDEEEKEYSTSSMQEQTD 390
Y K + + + + + ++K+ D+EE + S S + ++ D
Sbjct: 421 PPYEAKAAKRKAEYEKLIKAYDKKQASSADDEESDKSKSEVNDEDD 558
>TC229172 similar to UP|Q8IP68 (Q8IP68) CG31813-PA, partial (8%)
Length = 618
Score = 42.0 bits (97), Expect = 6e-04
Identities = 23/64 (35%), Positives = 42/64 (64%), Gaps = 4/64 (6%)
Frame = +1
Query: 424 VKTEEPCS---SSLSGGIDTVSDEGPDVDKKADEFIAKFREQIRLQRIESIKR-STRVAR 479
V+T P + + ++GG +++ +VD KAD+FI KF++Q++LQR++SI R + R
Sbjct: 328 VETRRPATVREAKVTGG---AAEDDAEVDAKADDFINKFKQQLKLQRLDSIIRYKEMIGR 498
Query: 480 NSSR 483
S++
Sbjct: 499 GSAK 510
>TC229173
Length = 924
Score = 41.6 bits (96), Expect = 8e-04
Identities = 21/53 (39%), Positives = 37/53 (69%), Gaps = 3/53 (5%)
Frame = +3
Query: 424 VKTEEPCS---SSLSGGIDTVSDEGPDVDKKADEFIAKFREQIRLQRIESIKR 473
V+T P + + ++GG +++ +VD KAD+FI KF++Q++LQR++SI R
Sbjct: 744 VETRRPATVREAKVTGG---AAEDDAEVDAKADDFINKFKQQLKLQRLDSIIR 893
>TC216023 similar to GB|AAM78036.1|21928015|AY125526 At2g01100/F23H14.7
{Arabidopsis thaliana;} , partial (15%)
Length = 1437
Score = 36.6 bits (83), Expect(2) = 9e-04
Identities = 51/208 (24%), Positives = 81/208 (38%), Gaps = 2/208 (0%)
Frame = +3
Query: 121 DEVDHQNQSESEDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKP 180
D DH+ + E K + WS+++ Y + +D +SSF E +
Sbjct: 429 DSGDHEKRKEKSSKRKTKKWSSESSDYSS------------------DDSESSFEEERRR 554
Query: 181 LLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTT--KRFSSNSFNRVRNYAEFEGLVEDKLK 238
+ L+ R S D + D LSK KR + F+ +Y+ EG D L
Sbjct: 555 KKKQSKKLRDRGSRSDSSNYDSTADELSKRKRQHKRQRPSKFSG-SDYSTDEG---DSLV 722
Query: 239 QKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKT 298
QK N + S +S S+ + + +A K K+ N
Sbjct: 723 QK--RNHGMHSKRHRQSESSESDL--SSDDAPHKKGHRKHHKHHKRSHNVELRLSDSDYR 890
Query: 299 SSIKFTPSESVAKNSEDLIKKKSFYYKS 326
S + + S+S+ +NSED KK+S KS
Sbjct: 891 SHGQRSRSKSLEENSEDESKKRSMDKKS 974
Score = 23.9 bits (50), Expect(2) = 9e-04
Identities = 8/10 (80%), Positives = 8/10 (80%)
Frame = +2
Query: 326 SYPPPPPPPP 335
S PPP PPPP
Sbjct: 992 SSPPPSPPPP 1021
>TC224762 homologue to UP|Q09085 (Q09085) Hydroxyproline-rich glycoprotein
(HRGP) (Fragment), partial (38%)
Length = 422
Score = 39.7 bits (91), Expect = 0.003
Identities = 15/22 (68%), Positives = 17/22 (77%)
Frame = +2
Query: 321 SFYYKSYPPPPPPPPPTMFYKS 342
S+YYKS PPP P PPP +YKS
Sbjct: 62 SYYYKSPPPPDPSPPPPYYYKS 127
Score = 38.9 bits (89), Expect = 0.005
Identities = 14/21 (66%), Positives = 17/21 (80%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP+ +YKS
Sbjct: 17 YYYKSPPPPDPSPPPSYYYKS 79
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 305 YYYKSPPPPSPSPPPPYYYKS 367
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 113 YYYKSPPPPSPSPPPPYYYKS 175
Score = 37.7 bits (86), Expect = 0.012
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 161 YYYKSPPPPSPSPPPHYYYKS 223
Score = 37.0 bits (84), Expect = 0.020
Identities = 13/21 (61%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YY+S PPP P PPP +YKS
Sbjct: 257 YYYRSPPPPSPSPPPPYYYKS 319
Score = 37.0 bits (84), Expect = 0.020
Identities = 13/21 (61%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +Y+S
Sbjct: 209 YYYKSPPPPDPSPPPPYYYRS 271
Score = 31.2 bits (69), Expect = 1.1
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P P +YKS
Sbjct: 353 YYYKSPPPPSPVSHPPYYYKS 415
>BE659924
Length = 588
Score = 39.3 bits (90), Expect = 0.004
Identities = 26/92 (28%), Positives = 36/92 (38%), Gaps = 8/92 (8%)
Frame = +2
Query: 327 YPPPPPP-------PPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEK-EDEDEEEK 378
YPPPPPP P Y T S Y K ++ TN + D D E
Sbjct: 65 YPPPPPPASDYFPEQPQKFSYTQTQTPSHYSHKTQTPSHYSHKTQQRTNPQIHDTDSERS 244
Query: 379 EYSTSSMQEQTDKERFIEKRVILEAEETDSDE 410
EY + +T+ E+ + E ET+ +E
Sbjct: 245 EYDFFDGKLETENEKTDSHHLPEEYTETEREE 340
>BQ785893 similar to PIR|T10863|T108 extensin precursor - kidney bean,
partial (17%)
Length = 429
Score = 39.3 bits (90), Expect = 0.004
Identities = 18/33 (54%), Positives = 19/33 (57%), Gaps = 9/33 (27%)
Frame = +3
Query: 318 KKKSFYYKSYPPPP---------PPPPPTMFYK 341
KKK +YY S PPPP PPPPP FYK
Sbjct: 102 KKKPYYYHSPPPPPPKKPYYYHSPPPPPKKFYK 200
Score = 29.3 bits (64), Expect = 4.1
Identities = 10/16 (62%), Positives = 11/16 (68%)
Frame = +3
Query: 325 KSYPPPPPPPPPTMFY 340
K PPPP PPPP +Y
Sbjct: 30 KHSPPPPSPPPPKPYY 77
>TC224734 homologue to UP|Q09085 (Q09085) Hydroxyproline-rich glycoprotein
(HRGP) (Fragment), partial (48%)
Length = 990
Score = 38.9 bits (89), Expect = 0.005
Identities = 18/41 (43%), Positives = 21/41 (50%)
Frame = +2
Query: 302 KFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKS 342
K P S + D +YYKS PPP P PPP +YKS
Sbjct: 50 KSPPPPSPSPPPPDPSPPPPYYYKSPPPPSPSPPPPYYYKS 172
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 302 YYYKSPPPPSPSPPPPYYYKS 364
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 206 YYYKSPPPPSPSPPPPYYYKS 268
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 254 YYYKSPPPPSPSPPPPYYYKS 316
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 158 YYYKSPPPPSPSPPPPYYYKS 220
Score = 33.1 bits (74), Expect = 0.28
Identities = 16/28 (57%), Positives = 18/28 (64%), Gaps = 7/28 (25%)
Frame = +2
Query: 322 FYYKSYPPP---PPP----PPPTMFYKS 342
+YYKS PPP PPP PPP +YKS
Sbjct: 41 YYYKSPPPPSPSPPPPDPSPPPPYYYKS 124
Score = 31.6 bits (70), Expect = 0.83
Identities = 12/18 (66%), Positives = 13/18 (71%)
Frame = +2
Query: 325 KSYPPPPPPPPPTMFYKS 342
KS PPP P PPP +YKS
Sbjct: 2 KSPPPPSPSPPPPYYYKS 55
Score = 31.2 bits (69), Expect = 1.1
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P P +YKS
Sbjct: 350 YYYKSPPPPSPVSHPPYYYKS 412
Score = 30.8 bits (68), Expect = 1.4
Identities = 12/21 (57%), Positives = 13/21 (61%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP PPP Y S
Sbjct: 398 YYYKSPPPPTSSPPPPYHYVS 460
>TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g47970
{Arabidopsis thaliana;} , partial (39%)
Length = 867
Score = 38.5 bits (88), Expect = 0.007
Identities = 28/104 (26%), Positives = 46/104 (43%), Gaps = 5/104 (4%)
Frame = +1
Query: 351 EKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSD- 409
E+ ++ G ++ N+ +DEDE+E++ + E I V E EE SD
Sbjct: 295 EEEESDVQRGGEPDDDDNDDDDEDEDEEDEEEQGEEADLGTEYLIRPLVTAEEEEASSDF 474
Query: 410 EDEDVGELKQ----SVKSVKTEEPCSSSLSGGIDTVSDEGPDVD 449
E E+ GE ++ K+E P S D+ D+G + D
Sbjct: 475 EPEENGEEEEEDVDDEDGEKSEAPPKRKRSDKDDSDDDDGGEDD 606
>AI441847 homologue to PIR|T11622|T11 extensin class 1 precursor - cowpea,
partial (29%)
Length = 429
Score = 38.5 bits (88), Expect = 0.007
Identities = 16/27 (59%), Positives = 19/27 (70%), Gaps = 5/27 (18%)
Frame = +1
Query: 322 FYYKSYPPPPP-----PPPPTMFYKST 343
+YYKS PPPPP PPPP +YKS+
Sbjct: 25 YYYKSPPPPPPYYYKSPPPPPYYYKSS 105
Score = 38.1 bits (87), Expect = 0.009
Identities = 16/26 (61%), Positives = 18/26 (68%), Gaps = 5/26 (19%)
Frame = +2
Query: 322 FYYKSYPPPPP-----PPPPTMFYKS 342
+YYKS PPPPP PPPP +YKS
Sbjct: 239 YYYKSPPPPPPYYYKSPPPPPYYYKS 316
Score = 33.5 bits (75), Expect = 0.22
Identities = 16/29 (55%), Positives = 19/29 (65%), Gaps = 8/29 (27%)
Frame = +2
Query: 322 FYYKSYPPPPP-----PPPPT---MFYKS 342
+YYKS PPPPP PPPP+ +YKS
Sbjct: 167 YYYKSPPPPPPYYYKSPPPPSPAPYYYKS 253
Score = 32.0 bits (71), Expect = 0.63
Identities = 15/32 (46%), Positives = 17/32 (52%), Gaps = 10/32 (31%)
Frame = +2
Query: 322 FYYKSYPPP----------PPPPPPTMFYKST 343
+YYKS PPP PPP P +YKST
Sbjct: 302 YYYKSPPPPSPTPYYYKSPPPPSPTPYYYKST 397
Score = 28.5 bits (62), Expect = 7.0
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPPP P +YKS
Sbjct: 128 YYYKS---PPPPSPTPYYYKS 181
>BQ741699 homologue to GP|3204132|emb extensin {Cicer arietinum}, partial
(69%)
Length = 431
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +3
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 12 YYYKSPPPPSPSPPPPYYYKS 74
Score = 37.7 bits (86), Expect = 0.012
Identities = 14/26 (53%), Positives = 17/26 (64%)
Frame = +3
Query: 317 IKKKSFYYKSYPPPPPPPPPTMFYKS 342
+ K +YY S PPP P PPP +YKS
Sbjct: 93 VPKPPYYYHSPPPPSPSPPPPYYYKS 170
Score = 35.0 bits (79), Expect = 0.075
Identities = 13/21 (61%), Positives = 15/21 (70%)
Frame = +3
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PP +YKS
Sbjct: 156 YYYKSPPPPSPSPPSLYYYKS 218
Score = 32.3 bits (72), Expect = 0.48
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = +3
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P P P +Y S
Sbjct: 60 YYYKSPPPPSPVPKPPYYYHS 122
Score = 29.6 bits (65), Expect = 3.1
Identities = 11/21 (52%), Positives = 13/21 (61%)
Frame = +3
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
++Y S PPP P PPP Y S
Sbjct: 249 YHYVSPPPPSPSPPPPYHYTS 311
Score = 29.6 bits (65), Expect = 3.1
Identities = 11/21 (52%), Positives = 13/21 (61%)
Frame = +3
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
++Y S PPP P P P YKS
Sbjct: 297 YHYTSPPPPSPAPAPKYIYKS 359
Score = 28.1 bits (61), Expect = 9.1
Identities = 10/15 (66%), Positives = 11/15 (72%)
Frame = +3
Query: 322 FYYKSYPPPPPPPPP 336
+YYKS PPP PPP
Sbjct: 204 YYYKSPPPPTSHPPP 248
>TC224765 similar to UP|Q09082 (Q09082) Extensin (Class I), partial (46%)
Length = 708
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +3
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 438 YYYKSPPPPSPSPPPPYYYKS 500
Score = 37.7 bits (86), Expect = 0.012
Identities = 14/26 (53%), Positives = 17/26 (64%)
Frame = +3
Query: 317 IKKKSFYYKSYPPPPPPPPPTMFYKS 342
+ K +YY S PPP P PPP +YKS
Sbjct: 375 VPKPPYYYHSPPPPSPSPPPPYYYKS 452
Score = 36.6 bits (83), Expect = 0.026
Identities = 15/34 (44%), Positives = 20/34 (58%)
Frame = +3
Query: 309 VAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKS 342
V K+ +K +YY+S PPP P P P +YKS
Sbjct: 159 VYKSPPPPVKSPPYYYQSPPPPSPKPKPPYYYKS 260
Score = 35.8 bits (81), Expect = 0.044
Identities = 13/21 (61%), Positives = 15/21 (70%)
Frame = +3
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +Y S
Sbjct: 486 YYYKSPPPPSPSPPPPYYYHS 548
Score = 34.7 bits (78), Expect = 0.097
Identities = 16/29 (55%), Positives = 17/29 (58%), Gaps = 8/29 (27%)
Frame = +3
Query: 322 FYYKSYPPP--------PPPPPPTMFYKS 342
+YYKS PPP PPPP PT YKS
Sbjct: 84 YYYKSPPPPSPVYIYKSPPPPSPTYVYKS 170
Score = 34.7 bits (78), Expect = 0.097
Identities = 13/24 (54%), Positives = 16/24 (66%)
Frame = +3
Query: 319 KKSFYYKSYPPPPPPPPPTMFYKS 342
K +YYKS PPP P P P +Y+S
Sbjct: 237 KPPYYYKSPPPPSPKPKPPYYYQS 308
Score = 34.3 bits (77), Expect = 0.13
Identities = 13/24 (54%), Positives = 16/24 (66%)
Frame = +3
Query: 319 KKSFYYKSYPPPPPPPPPTMFYKS 342
K +YY+S PPP P P P +YKS
Sbjct: 285 KPPYYYQSPPPPSPVPKPPYYYKS 356
Score = 34.3 bits (77), Expect = 0.13
Identities = 13/26 (50%), Positives = 16/26 (61%)
Frame = +3
Query: 317 IKKKSFYYKSYPPPPPPPPPTMFYKS 342
+ K +YYKS PPP P P P +Y S
Sbjct: 327 VPKPPYYYKSPPPPSPVPKPPYYYHS 404
Score = 32.7 bits (73), Expect = 0.37
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = +3
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YY S PPP P PP +YKS
Sbjct: 36 YYYHSPPPPSPSPPSPYYYKS 98
>TC208205 hydroxyproline-rich glycoprotein
Length = 603
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +1
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 259 YYYKSPPPPSPSPPPPYYYKS 321
Score = 33.9 bits (76), Expect = 0.17
Identities = 13/21 (61%), Positives = 14/21 (65%)
Frame = +1
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+ YKS PPP P PPP YKS
Sbjct: 19 YVYKSPPPPSPSPPPPYIYKS 81
Score = 33.5 bits (75), Expect = 0.22
Identities = 13/21 (61%), Positives = 14/21 (65%)
Frame = +1
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+ YKS PPP P PPP YKS
Sbjct: 115 YVYKSPPPPSPSPPPPYVYKS 177
Score = 33.5 bits (75), Expect = 0.22
Identities = 13/21 (61%), Positives = 14/21 (65%)
Frame = +1
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+ YKS PPP P PPP YKS
Sbjct: 67 YIYKSPPPPSPSPPPPYVYKS 129
Score = 33.5 bits (75), Expect = 0.22
Identities = 13/21 (61%), Positives = 14/21 (65%)
Frame = +1
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+ YKS PPP P PPP YKS
Sbjct: 163 YVYKSPPPPSPSPPPPYVYKS 225
Score = 32.0 bits (71), Expect = 0.63
Identities = 12/21 (57%), Positives = 14/21 (66%)
Frame = +1
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+ YKS PPP P PP +YKS
Sbjct: 211 YVYKSPPPPSPSPPSPYYYKS 273
Score = 31.6 bits (70), Expect = 0.83
Identities = 12/21 (57%), Positives = 13/21 (61%)
Frame = +1
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP PPP Y S
Sbjct: 307 YYYKSPPPPSHSPPPPYLYNS 369
>TC230101 similar to UP|Q41707 (Q41707) Extensin class 1 protein precursor,
partial (47%)
Length = 944
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 284 YYYKSPPPPSPSPPPPYYYKS 346
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 332 YYYKSPPPPSPSPPPPYYYKS 394
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 431 YYYKSPPPPSPSPPPPYYYKS 493
Score = 37.4 bits (85), Expect = 0.015
Identities = 13/21 (61%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYK+ PPP P PPP +YKS
Sbjct: 623 YYYKTAPPPSPSPPPPYYYKS 685
Score = 37.0 bits (84), Expect = 0.020
Identities = 13/21 (61%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YK+
Sbjct: 479 YYYKSPPPPSPSPPPPYYYKA 541
Score = 35.0 bits (79), Expect = 0.075
Identities = 13/21 (61%), Positives = 15/21 (70%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+ YKS PPP P PPP +YKS
Sbjct: 236 YLYKSPPPPSPSPPPPYYYKS 298
Score = 34.3 bits (77), Expect = 0.13
Identities = 12/22 (54%), Positives = 16/22 (72%)
Frame = +2
Query: 321 SFYYKSYPPPPPPPPPTMFYKS 342
++YYKS PPP P P P +YK+
Sbjct: 572 AYYYKSPPPPSPSPTPPYYYKT 637
Score = 34.3 bits (77), Expect = 0.13
Identities = 12/21 (57%), Positives = 15/21 (71%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYK+ PPP PPP +YKS
Sbjct: 527 YYYKAPPPPSASPPPAYYYKS 589
Score = 33.9 bits (76), Expect = 0.17
Identities = 12/22 (54%), Positives = 15/22 (67%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKST 343
+YYKS PPP PP PT +Y +
Sbjct: 671 YYYKSPPPPEKPPAPTPYYNKS 736
Score = 33.1 bits (74), Expect = 0.28
Identities = 16/40 (40%), Positives = 18/40 (45%), Gaps = 19/40 (47%)
Frame = +2
Query: 322 FYYKSYPPPP-------------------PPPPPTMFYKS 342
+YYKS PPPP PPPPP +YKS
Sbjct: 800 YYYKSAPPPPPYYYKSAPTPSAAPYYYESPPPPPPYYYKS 919
Score = 30.8 bits (68), Expect = 1.4
Identities = 14/29 (48%), Positives = 16/29 (54%), Gaps = 8/29 (27%)
Frame = +2
Query: 322 FYYKSYPPPPP--------PPPPTMFYKS 342
+YYKS P P P PPPP +YKS
Sbjct: 761 YYYKSPPTPSPTPYYYKSAPPPPPYYYKS 847
Score = 30.4 bits (67), Expect = 1.8
Identities = 11/19 (57%), Positives = 13/19 (67%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFY 340
+YYKS PPP P PT +Y
Sbjct: 380 YYYKSPPPPEKSPAPTPYY 436
Score = 30.4 bits (67), Expect = 1.8
Identities = 16/40 (40%), Positives = 20/40 (50%), Gaps = 5/40 (12%)
Frame = +2
Query: 308 SVAKNSEDLIKKKSFYYKSYPPPPP-----PPPPTMFYKS 342
++A S L+ + Y Y PPPP PPPP YKS
Sbjct: 131 AIALISITLVSVNADGYPYYSPPPPYQYTSPPPPPYLYKS 250
Score = 28.1 bits (61), Expect = 9.1
Identities = 24/109 (22%), Positives = 46/109 (42%), Gaps = 15/109 (13%)
Frame = -1
Query: 351 EKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQ-------EQTDKERFIEKRVILEA 403
E+ + + + + E N KE EEE+ Y + ++ +T R +E + + A
Sbjct: 818 EQTYSSTELERELVETCNSKELVMEEEETYCSKELELEAFQVVAETCNSRVVEGKEKVVA 639
Query: 404 EETDSDED--------EDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDE 444
+ +S E + V+ + EEPC+S + G + V +E
Sbjct: 638 QSCNSKVV*GMVKVVVETCNNKQVEVRQMVEEEPCNSKVGGVREMVVEE 492
>TC224645 homologue to UP|Q41707 (Q41707) Extensin class 1 protein precursor,
partial (58%)
Length = 856
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 755 YYYKSPPPPSPSPPPPYYYKS 817
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 179 YYYKSPPPPSPSPPPPYYYKS 241
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 227 YYYKSPPPPSPSPPPPYYYKS 289
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 275 YYYKSPPPPSPSPPPPYYYKS 337
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 323 YYYKSPPPPSPSPPPPYYYKS 385
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 371 YYYKSPPPPSPSPPPPYYYKS 433
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 419 YYYKSPPPPSPSPPPPYYYKS 481
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 467 YYYKSPPPPSPSPPPPYYYKS 529
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 515 YYYKSPPPPDPSPPPPYYYKS 577
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 563 YYYKSPPPPSPSPPPPYYYKS 625
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 611 YYYKSPPPPSPSPPPPYYYKS 673
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 659 YYYKSPPPPSPSPPPPYYYKS 721
Score = 38.1 bits (87), Expect = 0.009
Identities = 14/21 (66%), Positives = 16/21 (75%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PPP +YKS
Sbjct: 707 YYYKSPPPPSPSPPPPYYYKS 769
Score = 35.4 bits (80), Expect = 0.057
Identities = 15/22 (68%), Positives = 17/22 (77%)
Frame = +2
Query: 321 SFYYKSYPPPPPPPPPTMFYKS 342
S+YYKS PPP P PPP +YKS
Sbjct: 32 SYYYKSPPPPSPSPPP-YYYKS 94
Score = 35.0 bits (79), Expect = 0.075
Identities = 13/21 (61%), Positives = 15/21 (70%)
Frame = +2
Query: 322 FYYKSYPPPPPPPPPTMFYKS 342
+YYKS PPP P PP +YKS
Sbjct: 80 YYYKSPPPPSPSPPSPYYYKS 142
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.309 0.126 0.342
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,179,912
Number of Sequences: 63676
Number of extensions: 321160
Number of successful extensions: 13535
Number of sequences better than 10.0: 458
Number of HSP's better than 10.0 without gapping: 4483
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8895
length of query: 483
length of database: 12,639,632
effective HSP length: 101
effective length of query: 382
effective length of database: 6,208,356
effective search space: 2371591992
effective search space used: 2371591992
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC149079.6


