

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149079.10 - phase: 0
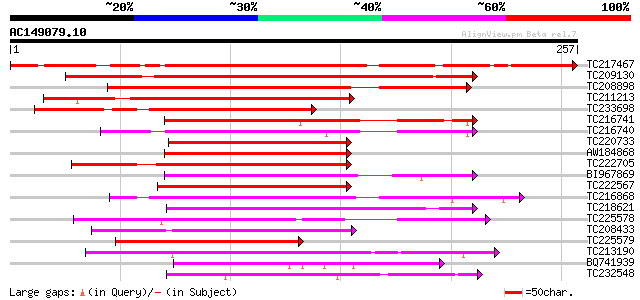
(257 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC217467 similar to UP|Q949B2 (Q949B2) AP2 domain-containing pro... 292 1e-79
TC209130 similar to PIR|T48518|T48518 transcription factor like ... 162 1e-40
TC208898 similar to PIR|T48518|T48518 transcription factor like ... 151 3e-37
TC211213 similar to PIR|T48518|T48518 transcription factor like ... 151 3e-37
TC233698 similar to PIR|T48518|T48518 transcription factor like ... 132 2e-31
TC216741 similar to UP|Q75UJ6 (Q75UJ6) DREB-like protein, partia... 122 2e-28
TC216740 similar to UP|O60585 (O60585) Ser/Arg-related nuclear m... 120 6e-28
TC220733 similar to GB|AAS68114.1|45592920|BT011790 At1g71450 {A... 119 1e-27
AW184868 119 2e-27
TC222705 similar to UP|Q6UK18 (Q6UK18) CaCBF1B, partial (54%) 117 5e-27
BI967869 similar to GP|12003384|gb Avr9/Cf-9 rapidly elicited pr... 108 2e-24
TC222567 weakly similar to UP|Q75UJ6 (Q75UJ6) DREB-like protein,... 108 3e-24
TC216868 homologue to UP|Q7Y0Y9 (Q7Y0Y9) Dehydration responsive ... 104 3e-23
TC218621 similar to UP|Q6UK18 (Q6UK18) CaCBF1B, partial (45%) 104 3e-23
TC225578 similar to UP|AP23_ARATH (P42736) AP2 domain transcript... 103 6e-23
TC208433 homologue to UP|Q9FH94 (Q9FH94) TINY-like protein, part... 99 2e-21
TC225579 similar to UP|Q84KS7 (Q84KS7) AP2/ERF-domain protein, p... 98 4e-21
TC213190 homologue to UP|Q7Y0Y9 (Q7Y0Y9) Dehydration responsive ... 97 6e-21
BQ741939 96 2e-20
TC232548 similar to UP|Q6R4U4 (Q6R4U4) DREB1, partial (43%) 94 6e-20
>TC217467 similar to UP|Q949B2 (Q949B2) AP2 domain-containing protein Rap211,
partial (48%)
Length = 1166
Score = 292 bits (747), Expect = 1e-79
Identities = 169/257 (65%), Positives = 195/257 (75%)
Frame = +2
Query: 1 MDREDSVLIHNSLCPQPTTSLSSYIIASTSSNSSSSLEEEEEEEEATNTKCKEKKKKKVG 60
M E+S + N + QPTTS SS+ I + S++SSSS+EE ATNT KEKKKK+
Sbjct: 62 MVMEESNPLDNEV--QPTTSSSSFTITTPSTSSSSSIEE------ATNTTTKEKKKKRA- 214
Query: 61 MVNKVNDRKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTI 120
++ + + +PTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTI
Sbjct: 215 -ISNIEGK--HPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTI 385
Query: 121 KGSSAYLNFPELAAVLPRAASASPKDIQVAAAKAAATVYNHAITDQIELEAEAEPSQAVS 180
KGSSAYLNFPELAA LPR AS SPKDIQ AAAKA+A + H EAE+E SQAVS
Sbjct: 386 KGSSAYLNFPELAAKLPRPASTSPKDIQAAAAKASALDFGHQ-----SHEAESELSQAVS 550
Query: 181 SSSSSSSSSSSSHNEGSSLKGEDDMFLNLPDLTLDLRHSGDDGFYYSSSAWLVNGAQHID 240
SS + SSSSSS+ SSLKG DD FL+LPDL+LDL H G D F+Y SSAWLV GA+HI+
Sbjct: 551 SSIQTQSSSSSSN---SSLKGVDDTFLDLPDLSLDLSH-GADEFHY-SSAWLVAGAEHIE 715
Query: 241 SSFRLEDPILWDSYQVT 257
FRLE+P LW+SY VT
Sbjct: 716 LGFRLEEPFLWESY*VT 766
>TC209130 similar to PIR|T48518|T48518 transcription factor like protein -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(45%)
Length = 1220
Score = 162 bits (410), Expect = 1e-40
Identities = 96/187 (51%), Positives = 117/187 (62%)
Frame = +3
Query: 26 IASTSSNSSSSLEEEEEEEEATNTKCKEKKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKW 85
+ S+ ++SSS+ + EA +K+ K+ + +P YRGVRMR WGKW
Sbjct: 219 LPSSEASSSSASSRKHSSSEAHLPAPDQKRPKQAR-----DSSSKHPVYRGVRMRAWGKW 383
Query: 86 VSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFPELAAVLPRAASASPK 145
VSEIREPRKK+RIWLGTF T +MAARAHDVAAL IKG+SA LNFPELAA LPR S SP+
Sbjct: 384 VSEIREPRKKNRIWLGTFATAEMAARAHDVAALAIKGNSAILNFPELAASLPRPDSNSPR 563
Query: 146 DIQVAAAKAAATVYNHAITDQIELEAEAEPSQAVSSSSSSSSSSSSSHNEGSSLKGEDDM 205
D+Q AAAKAAA N T + A S + SSS SS + SSSS +G S E
Sbjct: 564 DVQAAAAKAAAMEVNDVPTTPSSSSSSASLSLSQSSSWSSLAVSSSS-GDGPSTPEELGE 740
Query: 206 FLNLPDL 212
+ LP L
Sbjct: 741 IVELPSL 761
>TC208898 similar to PIR|T48518|T48518 transcription factor like protein -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(39%)
Length = 931
Score = 151 bits (381), Expect = 3e-37
Identities = 85/165 (51%), Positives = 103/165 (61%)
Frame = +3
Query: 45 EATNTKCKEKKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFP 104
E T+ + KK K++ + +P YRGVRMR WGKWVSEIREPRKKSRIWLGTFP
Sbjct: 255 EKLETEPESKKIKRIRGGGGGDSSNKHPLYRGVRMRNWGKWVSEIREPRKKSRIWLGTFP 434
Query: 105 TPDMAARAHDVAALTIKGSSAYLNFPELAAVLPRAASASPKDIQVAAAKAAATVYNHAIT 164
TP+MAARAHDVAAL+IKGS+A LNFP A LPR AS +P+D+Q AAAKAA
Sbjct: 435 TPEMAARAHDVAALSIKGSAAILNFPHFANSLPRPASLAPRDVQAAAAKAA--------- 587
Query: 165 DQIELEAEAEPSQAVSSSSSSSSSSSSSHNEGSSLKGEDDMFLNL 209
++ + S + SS+S S E SL+ DD F L
Sbjct: 588 ---HMDPSSLSSLVSAMDLSSASDELSQIIELPSLESTDDGFFVL 713
>TC211213 similar to PIR|T48518|T48518 transcription factor like protein -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(45%)
Length = 789
Score = 151 bits (381), Expect = 3e-37
Identities = 85/149 (57%), Positives = 100/149 (67%), Gaps = 8/149 (5%)
Frame = +2
Query: 16 QPTTSLSSYIIAST--------SSNSSSSLEEEEEEEEATNTKCKEKKKKKVGMVNKVND 67
+P + SS + ST S +SSSSL +++ KK+ N N+
Sbjct: 101 EPAVAASSELSDSTRNTNTNTPSPSSSSSLSPSPSPSSSSS------KKRARDNNNSRNN 262
Query: 68 RKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYL 127
+ YRGVRMR WGKWVSEIREPRKK+RIWLGTF T +MAARAHDVAALTIKGSSA L
Sbjct: 263 SNKHSVYRGVRMRTWGKWVSEIREPRKKNRIWLGTFATAEMAARAHDVAALTIKGSSAIL 442
Query: 128 NFPELAAVLPRAASASPKDIQVAAAKAAA 156
NFPELAA LPR AS SP+D+Q AAAKAA+
Sbjct: 443 NFPELAASLPRPASNSPRDVQAAAAKAAS 529
>TC233698 similar to PIR|T48518|T48518 transcription factor like protein -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(33%)
Length = 437
Score = 132 bits (331), Expect = 2e-31
Identities = 70/128 (54%), Positives = 88/128 (68%)
Frame = +3
Query: 12 SLCPQPTTSLSSYIIASTSSNSSSSLEEEEEEEEATNTKCKEKKKKKVGMVNKVNDRKNN 71
++ Q TTS + +S+SS+SSSS + + K + +K+ K + D +
Sbjct: 75 TMTQQATTSSETTTSSSSSSSSSSSTFSPSQH---SGPKAQAQKQSK-----RPRDCSKH 230
Query: 72 PTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFPE 131
P Y GVR R WGKWVSEIREPRKKSRIWLGTF TP+MAARAHDVAALTIKG +A LNFPE
Sbjct: 231 PVYHGVRKRNWGKWVSEIREPRKKSRIWLGTFATPEMAARAHDVAALTIKGEAAILNFPE 410
Query: 132 LAAVLPRA 139
+A +LPR+
Sbjct: 411 IADLLPRS 434
Score = 28.1 bits (61), Expect = 4.1
Identities = 15/27 (55%), Positives = 19/27 (69%)
Frame = +3
Query: 166 QIELEAEAEPSQAVSSSSSSSSSSSSS 192
QI + +A S ++SSSSSSSSSSS
Sbjct: 69 QITMTQQATTSSETTTSSSSSSSSSSS 149
Score = 26.9 bits (58), Expect = 9.2
Identities = 14/32 (43%), Positives = 19/32 (58%)
Frame = +3
Query: 163 ITDQIELEAEAEPSQAVSSSSSSSSSSSSSHN 194
+T Q +E S + SSSSSSS+ S S H+
Sbjct: 78 MTQQATTSSETTTSSSSSSSSSSSTFSPSQHS 173
>TC216741 similar to UP|Q75UJ6 (Q75UJ6) DREB-like protein, partial (36%)
Length = 769
Score = 122 bits (306), Expect = 2e-28
Identities = 72/145 (49%), Positives = 92/145 (62%), Gaps = 3/145 (2%)
Frame = +3
Query: 71 NPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFP 130
+P +RGVR R+WGKWVSEIREPRKKSRIWLG+FP P+MAA+A+DVAA +KG A LNFP
Sbjct: 150 HPLFRGVRKRRWGKWVSEIREPRKKSRIWLGSFPAPEMAAKAYDVAAYCLKGCKAQLNFP 329
Query: 131 -ELAAVLPRAASASPKDIQVAAAKAAATVYNHAITDQIELEAEAEPSQAVSSSSSSSSSS 189
E+ + P +S + +DIQ AAAKAA + QA S+ S SSS
Sbjct: 330 DEVHRLPPLPSSCTARDIQAAAAKAAHMMM----------------VQAASADSPEKSSS 461
Query: 190 SSSHNEGSSLKGEDDMF--LNLPDL 212
+S +GS G DD + + LP+L
Sbjct: 462 ITSDCDGS---GGDDFWGEIELPEL 527
>TC216740 similar to UP|O60585 (O60585) Ser/Arg-related nuclear matrix
protein, partial (3%)
Length = 800
Score = 120 bits (301), Expect = 6e-28
Identities = 76/174 (43%), Positives = 99/174 (56%), Gaps = 3/174 (1%)
Frame = +1
Query: 42 EEEEATNTKCKEKKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLG 101
++E AT T +K G + +P +RGVR R+WGKWVSEIREPRKKSRIWLG
Sbjct: 61 DDEAATTTSSGDKPATPRGGGTR------HPLFRGVRKRRWGKWVSEIREPRKKSRIWLG 222
Query: 102 TFPTPDMAARAHDVAALTIKGSSAYLNFPELAAVLPRAASA-SPKDIQVAAAKAAATVYN 160
+FP P+MAA+A+DVAA +KG A LNFP+ LP SA + +DIQ AAAKAA +
Sbjct: 223 SFPAPEMAAKAYDVAAYCLKGRKAQLNFPDEVHRLPLLPSACTARDIQAAAAKAAHMMI- 399
Query: 161 HAITDQIELEAEAEPSQAVSSSSSSSSSSSSSHNEGSSLKGEDDMF--LNLPDL 212
QA S ++ S SSS ++ G DD + + LP+L
Sbjct: 400 ---------------VQAASVTADSPEKSSSITSDCGDGGGGDDFWGEIELPEL 516
>TC220733 similar to GB|AAS68114.1|45592920|BT011790 At1g71450 {Arabidopsis
thaliana;} , partial (50%)
Length = 773
Score = 119 bits (299), Expect = 1e-27
Identities = 55/83 (66%), Positives = 69/83 (82%)
Frame = +1
Query: 73 TYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFPEL 132
+YRGVR R+WGKWVSEIREP KKSRIWLG++ +P+MAA A+DVAAL ++G +A LNFPEL
Sbjct: 91 SYRGVRQRKWGKWVSEIREPGKKSRIWLGSYESPEMAAAAYDVAALHLRGRAARLNFPEL 270
Query: 133 AAVLPRAASASPKDIQVAAAKAA 155
LPR S+ P+D+QVAA +AA
Sbjct: 271 VETLPRPTSSKPEDVQVAAQQAA 339
>AW184868
Length = 449
Score = 119 bits (297), Expect = 2e-27
Identities = 56/85 (65%), Positives = 68/85 (79%)
Frame = +1
Query: 71 NPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFP 130
+P YRGVR R+WGKWVSEIREPRKK+RIWLG+FP P+MAARA+DVAA +KG A LNFP
Sbjct: 67 HPMYRGVRKRRWGKWVSEIREPRKKNRIWLGSFPVPEMAARAYDVAAYCLKGRKAQLNFP 246
Query: 131 ELAAVLPRAASASPKDIQVAAAKAA 155
+ LP +S + +DIQ AAA+AA
Sbjct: 247 DDVDSLPLPSSRTARDIQTAAAQAA 321
>TC222705 similar to UP|Q6UK18 (Q6UK18) CaCBF1B, partial (54%)
Length = 745
Score = 117 bits (293), Expect = 5e-27
Identities = 64/127 (50%), Positives = 83/127 (64%)
Frame = +3
Query: 29 TSSNSSSSLEEEEEEEEATNTKCKEKKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVSE 88
+ +S +L +E+ A+N K + +KK +P YRGVR R GKWV E
Sbjct: 147 SGGSSRPALSDEDFFLAASNPKKRAGRKKF--------RETRHPVYRGVRRRDSGKWVCE 302
Query: 89 IREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFPELAAVLPRAASASPKDIQ 148
+REP KKSRIWLGTFPT +MAARAHDVAA+ ++G SA LNF + A+ LP A+A +DIQ
Sbjct: 303 VREPNKKSRIWLGTFPTAEMAARAHDVAAIALRGRSACLNFADSASRLPVPATAEARDIQ 482
Query: 149 VAAAKAA 155
AAA+AA
Sbjct: 483 KAAAEAA 503
>BI967869 similar to GP|12003384|gb Avr9/Cf-9 rapidly elicited protein 111B
{Nicotiana tabacum}, partial (47%)
Length = 743
Score = 108 bits (271), Expect = 2e-24
Identities = 60/149 (40%), Positives = 86/149 (57%), Gaps = 7/149 (4%)
Frame = -3
Query: 71 NPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFP 130
+P YRGVR R KWVSE+REP KK+RIWLGTFPTP+ ARAHDV A+ ++G A LNF
Sbjct: 699 HPVYRGVRRRNTDKWVSEVREPXKKTRIWLGTFPTPEXXARAHDVXAMALRGRYACLNFA 520
Query: 131 ELAAVLPRAASASPKDIQVAAAKAAATVYNHAITDQIELEAEAEPSQAVSSSSSS----- 185
+ LP A+A+ KDIQ AAA+AA PSQ + ++++
Sbjct: 519 DSTWRLPIPATANAKDIQKAAAEAAEAF---------------RPSQTLENTNTKQECVK 385
Query: 186 --SSSSSSSHNEGSSLKGEDDMFLNLPDL 212
++++ + G E++ L++P+L
Sbjct: 384 VVTTTTITEQKRGMFYTEEEEQVLDMPEL 298
>TC222567 weakly similar to UP|Q75UJ6 (Q75UJ6) DREB-like protein, partial
(35%)
Length = 580
Score = 108 bits (269), Expect = 3e-24
Identities = 53/88 (60%), Positives = 68/88 (77%)
Frame = +3
Query: 68 RKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYL 127
RK +RGVR R WG++VSEIR P +K+RIWLG+F +P+MAARA+D AA +KG+SA L
Sbjct: 21 RKRQSAFRGVRKRSWGRYVSEIRLPGQKTRIWLGSFGSPEMAARAYDSAAFFLKGTSATL 200
Query: 128 NFPELAAVLPRAASASPKDIQVAAAKAA 155
NFP+L LPR S+S +DIQ AAA+AA
Sbjct: 201 NFPDLVHSLPRPLSSSRRDIQSAAAEAA 284
>TC216868 homologue to UP|Q7Y0Y9 (Q7Y0Y9) Dehydration responsive element
binding protein, partial (53%)
Length = 1229
Score = 104 bits (260), Expect = 3e-23
Identities = 70/200 (35%), Positives = 101/200 (50%), Gaps = 12/200 (6%)
Frame = +3
Query: 46 ATNTKCKEKKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPT 105
++NT K +K+ K ++ YRG+RMR+WGKWV+EIREP K+SRIWLG++ T
Sbjct: 210 SSNTTITRKSEKR-----KQQHQQQEKPYRGIRMRKWGKWVAEIREPNKRSRIWLGSYAT 374
Query: 106 PDMAARAHDVAALTIKGSSAYLNFPELAAVLPRAASASPKDIQVAAAKAAATVYNHAITD 165
P AARA+D A ++G SA LNFPEL + ++ ++ + + AT
Sbjct: 375 PVAAARAYDTAVFHLRGPSARLNFPELLSQDDDVSTQQQGNMSADSIRKKAT-------- 530
Query: 166 QIELEAEAEPSQAVSSSSSSSSSSSSSHNEGSSL-------KGEDDMFLNLPDLTLDLRH 218
++ A + Q SSSS S++SSH S K ED F + + +D
Sbjct: 531 --QVGARVDALQTALQQSSSSHSANSSHVSSSDKPDLNEYPKPED**FPSTFKINIDPPI 704
Query: 219 SGDD-----GFYYSSSAWLV 233
G D GF SS L+
Sbjct: 705 MGSDVTYI*GFNLPSSPMLI 764
>TC218621 similar to UP|Q6UK18 (Q6UK18) CaCBF1B, partial (45%)
Length = 786
Score = 104 bits (260), Expect = 3e-23
Identities = 59/141 (41%), Positives = 80/141 (55%)
Frame = +3
Query: 72 PTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFPE 131
P YRGVR R KWV E+REP KK+RIWLG FP P+M ARAHDV A+ ++G A LNF +
Sbjct: 39 PVYRGVRRRNSDKWVCEVREPXKKTRIWLGIFPMPEMXARAHDVXAMALRGRYACLNFAD 218
Query: 132 LAAVLPRAASASPKDIQVAAAKAAATVYNHAITDQIELEAEAEPSQAVSSSSSSSSSSSS 191
A LP A A KDIQ AA +AA I E + AV+ ++ +++
Sbjct: 219 SAWRLPVPAMAEAKDIQKAAVEAA*AFRPDQILKNANTRQECVEAVAVAVVETTMATA-- 392
Query: 192 SHNEGSSLKGEDDMFLNLPDL 212
+G E++ L++P+L
Sbjct: 393 ---QGVFYMEEEEQVLDMPEL 446
>TC225578 similar to UP|AP23_ARATH (P42736) AP2 domain transcription factor
RAP2.3 (Related to AP2 protein 3) (Cadmium-induced
protein AS30), partial (38%)
Length = 1111
Score = 103 bits (258), Expect = 6e-23
Identities = 65/191 (34%), Positives = 97/191 (50%), Gaps = 2/191 (1%)
Frame = +3
Query: 30 SSNSSSSLEEEEEEEEATNTKCKEKKKKKVGMVNKVND--RKNNPTYRGVRMRQWGKWVS 87
++NS + ++ ++ + C++KKK VG K +D R YRG+R R WGKW +
Sbjct: 216 TTNSKNQPPLQKIPDKKVVSSCEKKKKSVVGAEKKKSDSGRARKNVYRGIRQRPWGKWAA 395
Query: 88 EIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFPELAAVLPRAASASPKDI 147
EIR+P K R+WLGTFPT + AA+A+D AA+ I+G A LNFP A + AA+ P
Sbjct: 396 EIRDPHKGVRVWLGTFPTAEEAAQAYDDAAIRIRGDKAKLNFP--ATTISAAAAPPPSKK 569
Query: 148 QVAAAKAAATVYNHAITDQIELEAEAEPSQAVSSSSSSSSSSSSSHNEGSSLKGEDDMFL 207
Q + ++ SSSSSS S++ + G S G D++ L
Sbjct: 570 QRCLS-----------------------PDIITEESSSSSSHSTTGSTGESGGGNDELDL 680
Query: 208 NLPDLTLDLRH 218
+ L L +
Sbjct: 681 KQIEWFLGLEN 713
>TC208433 homologue to UP|Q9FH94 (Q9FH94) TINY-like protein, partial (54%)
Length = 944
Score = 98.6 bits (244), Expect = 2e-21
Identities = 51/120 (42%), Positives = 72/120 (59%)
Frame = +3
Query: 38 EEEEEEEEATNTKCKEKKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVSEIREPRKKSR 97
E E E E + + + +++ KV + Y+G+RMR+WGKWV+EIREP K+SR
Sbjct: 195 EREREREMESEGRREGGERETTAATRKVEGAERR--YKGIRMRKWGKWVAEIREPNKRSR 368
Query: 98 IWLGTFPTPDMAARAHDVAALTIKGSSAYLNFPELAAVLPRAASASPKDIQVAAAKAAAT 157
IWLG++ TP AARA+D A ++G SA LNFPEL AA + D+ A+ + AT
Sbjct: 369 IWLGSYSTPVAAARAYDTAVFYLRGPSARLNFPELLVREGPAALVAGCDMSAASIRKKAT 548
>TC225579 similar to UP|Q84KS7 (Q84KS7) AP2/ERF-domain protein, partial (31%)
Length = 1156
Score = 97.8 bits (242), Expect = 4e-21
Identities = 45/85 (52%), Positives = 55/85 (63%)
Frame = +2
Query: 49 TKCKEKKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDM 108
+ C++KKKK V K R YRG+R R WGKW +EIR+P K R+WLGTFPT +
Sbjct: 302 SSCEKKKKKSVSAEKKSGGRARKNVYRGIRQRPWGKWAAEIRDPHKGVRVWLGTFPTAEE 481
Query: 109 AARAHDVAALTIKGSSAYLNFPELA 133
AARA+D AA I+G A LNFP A
Sbjct: 482 AARAYDDAAKRIRGDKAKLNFPATA 556
>TC213190 homologue to UP|Q7Y0Y9 (Q7Y0Y9) Dehydration responsive element
binding protein, partial (98%)
Length = 733
Score = 97.4 bits (241), Expect = 6e-21
Identities = 67/203 (33%), Positives = 98/203 (48%), Gaps = 15/203 (7%)
Frame = +3
Query: 35 SSLEEEEEEEEATNTKCKEKKKKKVGMVNKVNDRKNNP---------TYRGVRMRQWGKW 85
+S+ EE + + N+ KKK + +D+ N YRG+RMR+WGKW
Sbjct: 27 NSIMEERDHRCSNNSTMITTTKKKTSRRSPTSDKLKNQHPCEKQAMKPYRGIRMRKWGKW 206
Query: 86 VSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFPELAAVLPRAASASPK 145
V+EIREP K+SRIWLG++ TP AARA+D A ++G +A LNFPEL +
Sbjct: 207 VAEIREPNKRSRIWLGSYTTPMAAARAYDTAVFYLRGPTARLNFPELLFQDDQEEGNDSV 386
Query: 146 DIQVAAAKAAATVYNHAITDQIELEAEAEPSQAVSSSSSSSSSSSSSHNEGSSLKGEDD- 204
AA +A T Q+ +A + A+ +SS++S NE L+ D
Sbjct: 387 QHGAAAGNMSADSIRRKAT-QVGARVDALQT-ALHHHASSTNSLKPDLNEFPKLESLQD* 560
Query: 205 -----MFLNLPDLTLDLRHSGDD 222
FL+ + + GDD
Sbjct: 561 YRSIFNFLSPISIYYSFMYIGDD 629
>BQ741939
Length = 432
Score = 95.9 bits (237), Expect = 2e-20
Identities = 61/138 (44%), Positives = 78/138 (56%), Gaps = 15/138 (10%)
Frame = +2
Query: 75 RGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSA-YLNFPE-- 131
+GVRMR WG WVSEIR P +K+RIWLG++ T + AARA+D A L +KGSSA LNFP
Sbjct: 8 KGVRMRSWGSWVSEIRAPNQKTRIWLGSYSTAEAAARAYDAALLCLKGSSATNLNFPSSS 187
Query: 132 ---LAAVLPRAAS-ASPKDIQVAAAKA--------AATVYNHAITDQIELEAEAEPSQAV 179
L ++P+ S SPK IQ AA A A + N+ T + + P +
Sbjct: 188 SSLLLYIIPQDTSMMSPKSIQRVAAAAANNFLDNNAIAINNNVTTPPSPISPHSPPLAST 367
Query: 180 SSSSSSSSSSSSSHNEGS 197
SSSS SS S S + S
Sbjct: 368 PSSSSFVSSPSMSSSSPS 421
>TC232548 similar to UP|Q6R4U4 (Q6R4U4) DREB1, partial (43%)
Length = 800
Score = 94.0 bits (232), Expect = 6e-20
Identities = 61/145 (42%), Positives = 84/145 (57%), Gaps = 2/145 (1%)
Frame = -2
Query: 72 PTYRGVRMRQWGKWVSEIREPRKKS-RIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFP 130
P YRGVR R K E+R P KS RIWLGT+P P+ ARAHDVAAL ++G SA LNF
Sbjct: 793 PVYRGVRRRXXXKXXCEVRVPNDKSTRIWLGTYPVPEXXARAHDVAALALRGKSACLNFA 614
Query: 131 ELAAVLPRAASASPKDI-QVAAAKAAATVYNHAITDQIELEAEAEPSQAVSSSSSSSSSS 189
+ A LP AS + K+I +VAAA A A + +Q++ + + AV+ S S
Sbjct: 613 DSAWRLPLPASTNAKEIRRVAAAAAVAIAAEDSCGEQLQ---NSIVNDAVADDCEVSRSD 443
Query: 190 SSSHNEGSSLKGEDDMFLNLPDLTL 214
S + +S KG +F +L ++T+
Sbjct: 442 VSFDEDSNSNKGL-RVFCDLDEITM 371
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.308 0.122 0.343
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,851,525
Number of Sequences: 63676
Number of extensions: 222388
Number of successful extensions: 6626
Number of sequences better than 10.0: 371
Number of HSP's better than 10.0 without gapping: 2856
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4613
length of query: 257
length of database: 12,639,632
effective HSP length: 95
effective length of query: 162
effective length of database: 6,590,412
effective search space: 1067646744
effective search space used: 1067646744
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC149079.10


