

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148996.5 + phase: 0
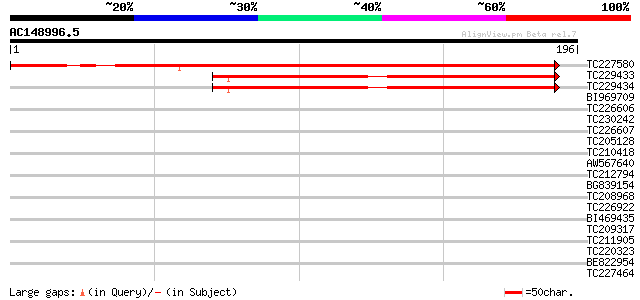
(196 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC227580 similar to PIR|S34045|S34045 protamine - North American... 226 5e-60
TC229433 similar to UP|SSB_ARATH (Q84J78) Single-stranded DNA-bi... 95 2e-20
TC229434 similar to UP|SSB_ARATH (Q84J78) Single-stranded DNA-bi... 89 1e-18
BI969709 37 0.006
TC226606 similar to UP|PIN1_MALDO (Q94G00) Peptidyl-prolyl cis-t... 35 0.029
TC230242 weakly similar to UP|Q9FQZ6 (Q9FQZ6) Avr9/Cf-9 rapidly ... 30 0.55
TC226607 similar to UP|PIN1_MALDO (Q94G00) Peptidyl-prolyl cis-t... 30 0.71
TC205128 similar to UP|Q9SMA5 (Q9SMA5) Zwh0004.1, partial (8%) 29 1.2
TC210418 similar to UP|Q9FMX6 (Q9FMX6) Gb|AAD21732.1, partial (36%) 29 1.6
AW567640 similar to SP|Q9C7E8|COL3_ Zinc finger protein constans... 29 1.6
TC212794 weakly similar to GB|AAQ55274.1|33942043|BT010323 At1g2... 29 1.6
BG839154 28 2.1
TC208968 similar to UP|GBF4_ARATH (P42777) G-box binding factor ... 28 2.1
TC226922 similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial (54%) 28 2.1
BI469435 28 2.1
TC209317 similar to UP|Q944G2 (Q944G2) Mevalonate kinase, partia... 28 2.7
TC211905 homologue to UP|Q77IW3 (Q77IW3) ORF91, partial (11%) 28 2.7
TC220323 similar to UP|LBD1_ARATH (Q9LQR0) LOB domain protein 1,... 28 2.7
BE822954 similar to GP|16417946|gb mevalonate kinase {Hevea bras... 28 2.7
TC227464 similar to UP|Q8VYI9 (Q8VYI9) AT5g64080/MHJ24_6, partia... 28 3.5
>TC227580 similar to PIR|S34045|S34045 protamine - North American opossum
{Didelphis virginiana;} , partial (31%)
Length = 930
Score = 226 bits (576), Expect = 5e-60
Identities = 116/193 (60%), Positives = 145/193 (75%), Gaps = 3/193 (1%)
Frame = +3
Query: 1 MAKFSISRTSTLYRSLLSTPPPPLHHHRHIPLRFCTTTTSLSDYDDADSAAPSPSPE--- 57
M+ ++ + LY SLL TP + +RH +T L D D+ADSA P+P+
Sbjct: 30 MSSSMMTLSRRLYCSLLRTP----NAYRHF------STDHLFDSDEADSAPDPPAPDAVQ 179
Query: 58 QTERTFFDRPLENGLDPGVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPL 117
+ E+ DRPLENGLD G+YRAILVGKAGQ PLQKKLKSGT VTLLS+GTGGI NNRRP
Sbjct: 180 EEEKFVIDRPLENGLDVGIYRAILVGKAGQAPLQKKLKSGTSVTLLSVGTGGIHNNRRPR 359
Query: 118 DHENPREYANRCAVQWHRVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVR 177
ENP++YANR A+QWHRV+VYP++LG+L+ K V+PGS LYVEGNLETK F+DP+TG+ R
Sbjct: 360 VDENPKDYANRSAIQWHRVSVYPQKLGDLVTKHVVPGSMLYVEGNLETKCFTDPITGIAR 539
Query: 178 RVREVAVRRNGNL 190
R+RE+AVRRNG +
Sbjct: 540 RIREIAVRRNGRI 578
>TC229433 similar to UP|SSB_ARATH (Q84J78) Single-stranded DNA-binding
protein, mitochondrial precursor, partial (73%)
Length = 846
Score = 94.7 bits (234), Expect = 2e-20
Identities = 46/127 (36%), Positives = 80/127 (62%), Gaps = 7/127 (5%)
Frame = +1
Query: 71 GLDP-------GVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPR 123
G+DP GV++AI+ GK GQ P+QK L++G +T+ ++GTGG+ + R + P+
Sbjct: 229 GVDPKKGWGFRGVHKAIICGKVGQAPVQKILRNGRNITIFTVGTGGMFDQRIQGPKDLPK 408
Query: 124 EYANRCAVQWHRVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVA 183
QWHR+ V+ + LG ++ + S++YVEG++ET+V++D + G V+ + E+
Sbjct: 409 P------AQWHRIAVHNDILGTYAVQQLFKNSSVYVEGDIETRVYNDSINGNVKSIPEIC 570
Query: 184 VRRNGNL 190
VRR+G +
Sbjct: 571 VRRDGRI 591
>TC229434 similar to UP|SSB_ARATH (Q84J78) Single-stranded DNA-binding
protein, mitochondrial precursor, partial (75%)
Length = 761
Score = 89.0 bits (219), Expect = 1e-18
Identities = 44/127 (34%), Positives = 78/127 (60%), Gaps = 7/127 (5%)
Frame = +2
Query: 71 GLDP-------GVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPR 123
G+DP GV++AI+ GK GQ P+QK L++G + + ++GTGG+ + R + P+
Sbjct: 65 GVDPKKGWGFRGVHKAIICGKVGQAPVQKILRNGRNLXIFTVGTGGMFDQRIQGPKDLPK 244
Query: 124 EYANRCAVQWHRVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREVA 183
QWHR+ V+ + LG ++ + S++YVEG++E +V++D + G V+ + E+
Sbjct: 245 P------AQWHRIAVHNDILGAYAVQKLFKNSSVYVEGDIEIRVYNDSINGEVKSIPEIC 406
Query: 184 VRRNGNL 190
VRR+G +
Sbjct: 407 VRRDGKI 427
>BI969709
Length = 572
Score = 37.0 bits (84), Expect = 0.006
Identities = 24/76 (31%), Positives = 35/76 (45%)
Frame = -3
Query: 120 ENPREYANRCAVQWHRVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRV 179
+ PR + N Q R+ + + LG + +YVEG E +VF D TG V+
Sbjct: 468 QGPRVFPN--LAQGLRIPGHNDILGPYPVHHFFKTFPVYVEGAFELRVFIDAFTGTVKTF 295
Query: 180 REVAVRRNGNLANSSN 195
++ V RNG S N
Sbjct: 294 PKICVPRNGRFPFSKN 247
>TC226606 similar to UP|PIN1_MALDO (Q94G00) Peptidyl-prolyl cis-trans
isomerase 1 (Rotamase Pin1) (PPIase Pin1) (MdPin1) ,
partial (91%)
Length = 748
Score = 34.7 bits (78), Expect = 0.029
Identities = 25/66 (37%), Positives = 32/66 (47%)
Frame = -1
Query: 7 SRTSTLYRSLLSTPPPPLHHHRHIPLRFCTTTTSLSDYDDADSAAPSPSPEQTERTFFDR 66
SRT LY+ + S P P HHH+ TTTTS + S +PSP P T + D
Sbjct: 196 SRTFVLYQYVRS--P*PHHHHQSST----TTTTSSTS*SYCGSPSPSPPPRSTPSSASDL 35
Query: 67 PLENGL 72
L + L
Sbjct: 34 TLSSPL 17
>TC230242 weakly similar to UP|Q9FQZ6 (Q9FQZ6) Avr9/Cf-9 rapidly elicited
protein 146, partial (30%)
Length = 713
Score = 30.4 bits (67), Expect = 0.55
Identities = 9/13 (69%), Positives = 11/13 (84%)
Frame = +1
Query: 19 TPPPPLHHHRHIP 31
+PPPPLHHH +P
Sbjct: 355 SPPPPLHHHHQVP 393
Score = 28.1 bits (61), Expect = 2.7
Identities = 14/47 (29%), Positives = 21/47 (43%)
Frame = +2
Query: 3 KFSISRTSTLYRSLLSTPPPPLHHHRHIPLRFCTTTTSLSDYDDADS 49
+FS S + L+ + + HHH CTTTT +DD +
Sbjct: 290 EFSCSNSPALFHVIKRSTNKHRHHHHR-----CTTTTKFPHHDDVST 415
>TC226607 similar to UP|PIN1_MALDO (Q94G00) Peptidyl-prolyl cis-trans
isomerase 1 (Rotamase Pin1) (PPIase Pin1) (MdPin1) ,
partial (93%)
Length = 736
Score = 30.0 bits (66), Expect = 0.71
Identities = 21/58 (36%), Positives = 27/58 (46%)
Frame = -3
Query: 15 SLLSTPPPPLHHHRHIPLRFCTTTTSLSDYDDADSAAPSPSPEQTERTFFDRPLENGL 72
S+ + P P HH TTTTS + Y A S +PSP P T + D L + L
Sbjct: 209 SICAKP*PQYRHHPST-----TTTTSSTSY--AYSGSPSPPPRSTPSSASDLTLSSPL 57
>TC205128 similar to UP|Q9SMA5 (Q9SMA5) Zwh0004.1, partial (8%)
Length = 411
Score = 29.3 bits (64), Expect = 1.2
Identities = 9/10 (90%), Positives = 9/10 (90%)
Frame = +2
Query: 20 PPPPLHHHRH 29
PPPPLHHH H
Sbjct: 113 PPPPLHHHHH 142
>TC210418 similar to UP|Q9FMX6 (Q9FMX6) Gb|AAD21732.1, partial (36%)
Length = 598
Score = 28.9 bits (63), Expect = 1.6
Identities = 13/22 (59%), Positives = 14/22 (63%)
Frame = +2
Query: 8 RTSTLYRSLLSTPPPPLHHHRH 29
+TST SL PPPPL H RH
Sbjct: 26 QTSTTSVSLSPMPPPPLSHARH 91
>AW567640 similar to SP|Q9C7E8|COL3_ Zinc finger protein constans-like 3.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (11%)
Length = 386
Score = 28.9 bits (63), Expect = 1.6
Identities = 18/58 (31%), Positives = 21/58 (36%), Gaps = 13/58 (22%)
Frame = +2
Query: 23 PLHHHRHIPLRFCTTTTSLSDYDDAD-------------SAAPSPSPEQTERTFFDRP 67
PL HH H P TT T Y A+ P+PSP T + F P
Sbjct: 125 PLTHHHHSPCCHATTATPNPPYSSAELIPLNSV*FATSTCTPPTPSPSSTCASRFATP 298
>TC212794 weakly similar to GB|AAQ55274.1|33942043|BT010323 At1g23980
{Arabidopsis thaliana;} , partial (7%)
Length = 625
Score = 28.9 bits (63), Expect = 1.6
Identities = 16/49 (32%), Positives = 25/49 (50%), Gaps = 4/49 (8%)
Frame = -2
Query: 8 RTSTLYRSLLSTPPP----PLHHHRHIPLRFCTTTTSLSDYDDADSAAP 52
R+S L+ S LS+ PP P HHH H+ + T + S+++ P
Sbjct: 192 RSSHLWLSPLSSSPPHSHFPKHHHHHLEIENEITRLANSNHEPHTDTNP 46
>BG839154
Length = 787
Score = 28.5 bits (62), Expect = 2.1
Identities = 12/20 (60%), Positives = 13/20 (65%)
Frame = -2
Query: 9 TSTLYRSLLSTPPPPLHHHR 28
TS LY L S+PPP HH R
Sbjct: 597 TSHLYHYLSSSPPPHFHHSR 538
>TC208968 similar to UP|GBF4_ARATH (P42777) G-box binding factor 4
(AtbZIP40), partial (27%)
Length = 1199
Score = 28.5 bits (62), Expect = 2.1
Identities = 14/50 (28%), Positives = 27/50 (54%)
Frame = +2
Query: 20 PPPPLHHHRHIPLRFCTTTTSLSDYDDADSAAPSPSPEQTERTFFDRPLE 69
PPPP +P +++S+ + + SAAPS S ++ +R + P++
Sbjct: 377 PPPPPPPSSFLPFPADGSSSSVEPFANGVSAAPSNSVQKGKRRAVEEPVD 526
>TC226922 similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial (54%)
Length = 1190
Score = 28.5 bits (62), Expect = 2.1
Identities = 10/14 (71%), Positives = 10/14 (71%)
Frame = +3
Query: 20 PPPPLHHHRHIPLR 33
PPPP HHHR LR
Sbjct: 153 PPPPSHHHRQTSLR 194
>BI469435
Length = 426
Score = 28.5 bits (62), Expect = 2.1
Identities = 20/53 (37%), Positives = 24/53 (44%)
Frame = +3
Query: 10 STLYRSLLSTPPPPLHHHRHIPLRFCTTTTSLSDYDDADSAAPSPSPEQTERT 62
S+ LLS HH+ P TT+ S S A SA P PSP+ T T
Sbjct: 6 SSTLTPLLSPHASSSQHHKP-PAAATTTSLS*STSTAAPSAPPPPSPQTTTTT 161
>TC209317 similar to UP|Q944G2 (Q944G2) Mevalonate kinase, partial (38%)
Length = 534
Score = 28.1 bits (61), Expect = 2.7
Identities = 15/45 (33%), Positives = 22/45 (48%), Gaps = 4/45 (8%)
Frame = -2
Query: 16 LLSTPPPPL----HHHRHIPLRFCTTTTSLSDYDDADSAAPSPSP 56
L+STPPPP+ H + H T +T++ D + S P P
Sbjct: 425 LISTPPPPIPAMKHWNPHDSSSATTLSTTVPDSNVGSSVRTQPPP 291
>TC211905 homologue to UP|Q77IW3 (Q77IW3) ORF91, partial (11%)
Length = 639
Score = 28.1 bits (61), Expect = 2.7
Identities = 14/33 (42%), Positives = 16/33 (48%)
Frame = +2
Query: 8 RTSTLYRSLLSTPPPPLHHHRHIPLRFCTTTTS 40
RT L LL PPPPL H+ L C T+
Sbjct: 2 RTLILLSLLLPPPPPPLAFSHHLSLLCCVV*TA 100
>TC220323 similar to UP|LBD1_ARATH (Q9LQR0) LOB domain protein 1, partial
(69%)
Length = 854
Score = 28.1 bits (61), Expect = 2.7
Identities = 15/45 (33%), Positives = 21/45 (46%)
Frame = +1
Query: 14 RSLLSTPPPPLHHHRHIPLRFCTTTTSLSDYDDADSAAPSPSPEQ 58
++ STP PP+ H +P + S S + S P PSP Q
Sbjct: 88 KTTTSTPSPPIPIH--LPTMLSNSPPSSSSSPPSQSPTPFPSPSQ 216
>BE822954 similar to GP|16417946|gb mevalonate kinase {Hevea brasiliensis},
partial (34%)
Length = 481
Score = 28.1 bits (61), Expect = 2.7
Identities = 15/45 (33%), Positives = 22/45 (48%), Gaps = 4/45 (8%)
Frame = +2
Query: 16 LLSTPPPPL----HHHRHIPLRFCTTTTSLSDYDDADSAAPSPSP 56
L+STPPPP+ H + H T +T++ D + S P P
Sbjct: 92 LISTPPPPIPAMKHWNPHDSSSATTLSTTVPDSNVGSSVRTQPPP 226
>TC227464 similar to UP|Q8VYI9 (Q8VYI9) AT5g64080/MHJ24_6, partial (53%)
Length = 829
Score = 27.7 bits (60), Expect = 3.5
Identities = 18/58 (31%), Positives = 27/58 (46%), Gaps = 11/58 (18%)
Frame = +3
Query: 22 PPLHHHRHIPLRF----CTTTTSLSDYDDADSAAPSPSPEQTER-------TFFDRPL 68
P HHH H PL T T S + A ++P+ +P Q++R + F +PL
Sbjct: 72 PREHHHHHHPLTRQRPPWTAPTWFSPWPIACPSSPTAAPSQSQRAHAALV*SLFSKPL 245
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.134 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,275,286
Number of Sequences: 63676
Number of extensions: 157604
Number of successful extensions: 2027
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 1829
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1978
length of query: 196
length of database: 12,639,632
effective HSP length: 92
effective length of query: 104
effective length of database: 6,781,440
effective search space: 705269760
effective search space used: 705269760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC148996.5


