

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
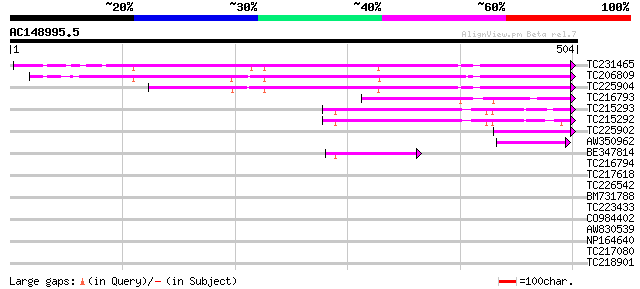
Query= AC148995.5 + phase: 0
(504 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC231465 UP|Q8GUS1 (Q8GUS1) NADPH:P450 reductase, partial (77%) 235 3e-62
TC206809 homologue to UP|NCPR_PHAAU (P37116) NADPH--cytochrome P... 211 5e-55
TC225904 similar to UP|O48937 (O48937) NADPH cytochrome P450 red... 210 1e-54
TC216793 similar to UP|FENS_PEA (Q41014) Ferredoxin--NADP reduct... 66 4e-11
TC215293 similar to UP|FENR_TOBAC (O04977) Ferredoxin--NADP redu... 59 4e-09
TC215292 similar to UP|FENR_PEA (P10933) Ferredoxin--NADP reduct... 57 1e-08
TC225902 similar to UP|O48937 (O48937) NADPH cytochrome P450 red... 51 1e-06
AW350962 homologue to GP|27261128|gb| NADPH:P450 reductase {Glyc... 47 2e-05
BE347814 similar to SP|P41346|FENR Ferredoxin--NADP reductase c... 45 8e-05
TC216794 homologue to UP|FENS_PEA (Q41014) Ferredoxin--NADP redu... 40 0.002
TC217618 similar to GB|AAO64868.1|29028978|BT005933 At5g50860 {A... 32 0.67
TC226542 weakly similar to UP|Q9ZWQ5 (Q9ZWQ5) UDP-glycose:flavon... 30 3.3
BM731788 30 3.3
TC223433 weakly similar to UP|Q9AST4 (Q9AST4) AT4g31930/F10N7_26... 29 5.6
CO984402 28 7.4
AW830539 similar to SP|O04450|TCPE_ T-complex protein 1 epsilon... 28 7.4
NP164640 receptor-like protein kinase 28 9.6
TC217080 similar to UP|Q94EJ2 (Q94EJ2) At1g08460/T27G7_7 (HDA8),... 28 9.6
TC218901 similar to UP|Q9LGX2 (Q9LGX2) ESTs AU082689(E31988), pa... 28 9.6
>TC231465 UP|Q8GUS1 (Q8GUS1) NADPH:P450 reductase, partial (77%)
Length = 1933
Score = 235 bits (600), Expect = 3e-62
Identities = 162/518 (31%), Positives = 252/518 (48%), Gaps = 18/518 (3%)
Frame = +1
Query: 4 VTKKLDKRLMDLGGKAILERGLGDDQQPSGYEGTLDPWLSSLWRMLNMIKPELLPSGPDV 63
+ K +D+ L + G K ++ GLGDD Q E W SLW L+ +LL DV
Sbjct: 97 IGKIVDEELSEQGAKRLVPLGLGDDDQ--SIEDDFVAWKESLWSELD----QLLRDEDDV 258
Query: 64 LIQDTTLIDQPKVQITYHNIENVVSHSSDTGSVRSMHPGKSSSNR--NGYPDCFLKLVKN 121
T+ K I + VV H S S H ++ N + + C + +
Sbjct: 259 ----NTVSTPYKAAIPEYR---VVIHDSTVTSCNDNHLNVANGNAVFDIHHPCRVNIAAQ 417
Query: 122 LPLTKPNSGKDVRHFEFEFVSHAIEYDTGDILEVLPGQDSAAVDAFIRRCNLDPDSLITV 181
L KP S + H EF+ I Y+TGD + V V+ + D D + ++
Sbjct: 418 RELHKPESDRSCIHLEFDISGTGIIYETGDHVGVFAENGDETVEEAGKLLGQDLDLVFSI 597
Query: 182 SPKGMDCNGHGSRMPVKLRTFVELTMDVASAS-----PRRYFFEARCSE----HERERLE 232
D GS +P L +A + PR+ A + E +RL
Sbjct: 598 HTNNEDGTPLGSSLPPPFPGPCTLRFALAHYADLLNPPRKASLVALAAHTSEPSEADRLT 777
Query: 233 YFASPEGRDDLYQYNQKERRTVLEVLKDFPSVQMPLE-WLIQLVPMLKKREFSISSSQSS 291
+ +SP+G+D+ ++ +R++LEV+ +FPS + PL + + P L+ R +SISSS
Sbjct: 778 FLSSPQGKDEYSKWLVGSQRSLLEVMAEFPSAKPPLGVFFAAVAPHLQPRYYSISSSPRF 957
Query: 292 HPNQVHLTVSVVSWTTPYKRKKKGLCSSWLAALDP----RDAVSLPVWFQKGSLPTPSP- 346
P +VH+T ++V TP R KG+CS+W+ P RD P++ + + P+
Sbjct: 958 SPQKVHVTCALVCGPTPTGRIHKGVCSTWMKNAIPLEKSRDCSWAPIFIRTSNFKLPADH 1137
Query: 347 SLPLILVGPGTGCAPFRGFIEER-ALQSKTISIAPIIFFFGCWNEDGDFLYKDFWLNHSQ 405
S+P+I+VGPGTG APFRGF++ER AL+ + + P + FFGC N DF+Y+D L +
Sbjct: 1138SIPIIMVGPGTGLAPFRGFLQERLALKEDAVQLGPALLFFGCRNRQMDFIYEDE-LKNFM 1314
Query: 406 NNGVLSESTGGGFYVAFSRDQPEKVYVQHKLREHSGRVWNLLAEGASVYIAGSLTKMPTD 465
G LSE V FSR+ PEK YVQHK+ + + +WNL+++G +Y+ G M D
Sbjct: 1315EQGALSE-----LIVTFSREGPEKEYVQHKMMDKAANLWNLISQGGYLYVCGDAKGMARD 1479
Query: 466 VTSAFEEIVSKENDVSKEDAVRWIRALEKCGKYHIEAW 503
V IV ++ +V A ++ L+ G+Y + W
Sbjct: 1480VHRTLHTIVQQQENVDSSKAEAIVKKLQMDGRYLRDVW 1593
>TC206809 homologue to UP|NCPR_PHAAU (P37116) NADPH--cytochrome P450
reductase (CPR) (P450R) , partial (70%)
Length = 1867
Score = 211 bits (538), Expect = 5e-55
Identities = 156/506 (30%), Positives = 244/506 (47%), Gaps = 20/506 (3%)
Frame = +2
Query: 18 KAILERGLGDDQQPSGYEGTLDPWLSSLWRMLNMIKPELLPSGPDVLIQDTTLIDQPKVQ 77
K ++ GLGDD Q E W +LW PEL D L++D +
Sbjct: 2 KRLVTFGLGDDDQ--SIEDDFSAWKETLW-------PEL-----DQLLRDEDDANTVSTP 139
Query: 78 ITYHNIE-NVVSHSSDTGSVRSMHPGKSSSNR--NGYPDCFLKLVKNLPLTKPNSGKDVR 134
T +E VV H S H ++ N + + C + + L KP S +
Sbjct: 140 YTAAILEYRVVIHDPTFTSSYDNHVTVANGNAVFDIHHPCRVNVAVQRELHKPESDRSCI 319
Query: 135 HFEFEFVSHAIEYDTGDILEVLPGQDSAAVDAFIRRCNLDPDSLITVSPKGMDCNGHGSR 194
H EF+ + Y+TGD + V V+ + + D L ++ D G
Sbjct: 320 HLEFDISGTGLTYETGDHVGVYADNCDETVEETGKLLGQNLDLLFSLHTDKEDGTSLGGS 499
Query: 195 M------PVKLRTFVELTMDVASASPRRYFFEARCSE----HERERLEYFASPEGRDDLY 244
+ P LRT + D+ + PR+ A S E ERL++ +SP+G+D+
Sbjct: 500 LLPPFPGPCTLRTALARYADLLNP-PRKAALVALASHASEPSEAERLKFLSSPQGKDEYS 676
Query: 245 QYNQKERRTVLEVLKDFPSVQMPLE-WLIQLVPMLKKREFSISSSQSSHPNQVHLTVSVV 303
++ +R++LEV+ +FPS + PL + + P L+ R +SISSS P +VH+T ++V
Sbjct: 677 KWVVGSQRSLLEVMAEFPSAKPPLGVFFAAVAPRLQPRYYSISSSPRFAPQRVHVTCALV 856
Query: 304 SWTTPYKRKKKGLCSSWLAALDPR----DAVSLPVWFQKGSLPTP-SPSLPLILVGPGTG 358
TP R KG+CS+W+ P D P++ + + P S+P+I+VGPGTG
Sbjct: 857 YGPTPTGRIHKGVCSTWMKNAIPLEKSPDCSWAPIFIRPSNFKLPVDHSIPIIMVGPGTG 1036
Query: 359 CAPFRGFIEER-ALQSKTISIAPIIFFFGCWNEDGDFLYKDFWLNHSQNNGVLSESTGGG 417
APFRGF++ER AL+ + P I FFGC N DF+Y++ N + G LSE
Sbjct: 1037LAPFRGFLQERFALKEAGVQQGPAILFFGCRNRRLDFIYEEELKNFVEQ-GSLSE----- 1198
Query: 418 FYVAFSRDQPEKVYVQHKLREHSGRVWNLLAEGASVYIAGSLTKMPTDVTSAFEEIVSKE 477
VAFSR+ EK YVQHK+ + + ++W+L+++G +Y+ G M DV IV ++
Sbjct: 1199LIVAFSREGAEKEYVQHKMMDQAAQLWSLISQGGYLYVCGDAKGMARDVHRTLHTIVQQQ 1378
Query: 478 NDVSKEDAVRWIRALEKCGKYHIEAW 503
+V A ++ L+ G+Y + W
Sbjct: 1379ENVDSTKAEAIVKKLQMDGRYLRDVW 1456
>TC225904 similar to UP|O48937 (O48937) NADPH cytochrome P450 reductase ,
partial (68%)
Length = 1704
Score = 210 bits (534), Expect = 1e-54
Identities = 130/397 (32%), Positives = 209/397 (51%), Gaps = 17/397 (4%)
Frame = +1
Query: 124 LTKPNSGKDVRHFEFEFVSHAIEYDTGDILEVLPGQDSAAVDAFIRRCNLDPDSLITVSP 183
L P S + H EF+ + Y+TGD + V S V+ IR L PD+ ++
Sbjct: 253 LHTPASDRSCTHLEFDISGTGVTYETGDHVGVYCENLSETVEEAIRLIGLSPDTYFSIHT 432
Query: 184 KGMDCNGH-GSRMP-----VKLRTFVELTMDVASASPRRYFFEARCSE----HERERLEY 233
D GS +P LR + DV S SP++ A + E +RL +
Sbjct: 433 DDEDGKPRSGSSLPPTFPPCTLRKALTQYADVLS-SPKKSALLALAAHASDPSEADRLRH 609
Query: 234 FASPEGRDDLYQYNQKERRTVLEVLKDFPSVQMPLE-WLIQLVPMLKKREFSISSSQSSH 292
ASP G+D+ ++ +R++LEV+ +FPS + P+ + + P L+ R +SISSS
Sbjct: 610 LASPAGKDEYSEWVIASQRSLLEVMAEFPSAKPPIGVFFAAVAPRLQPRFYSISSSPRMV 789
Query: 293 PNQVHLTVSVVSWTTPYKRKKKGLCSSWLAALDP----RDAVSLPVWFQKGSLPTPSPS- 347
PN++H+T ++V P R KG+CS+W+ P +D P++ + + PS +
Sbjct: 790 PNRIHVTCALVHEKMPTGRIHKGVCSTWMKNSVPLEKSQDCSWAPIFVRTSNFRLPSDNK 969
Query: 348 LPLILVGPGTGCAPFRGFIEER-ALQSKTISIAPIIFFFGCWNEDGDFLYKDFWLNHSQN 406
+P+I++GPGTG APFRGF++ER AL+ + P + FFGC N D++Y+D L+H N
Sbjct: 970 VPIIMIGPGTGLAPFRGFLQERLALKEGGAELGPSVLFFGCRNRQMDYIYEDE-LSHFVN 1146
Query: 407 NGVLSESTGGGFYVAFSRDQPEKVYVQHKLREHSGRVWNLLAEGASVYIAGSLTKMPTDV 466
G L E +AFSR+ P K YVQHK+ E + +W+++++GA +Y+ G M DV
Sbjct: 1147TGALDE-----LILAFSREGPTKEYVQHKMMEKASEIWSMISQGAYIYVCGDAKGMARDV 1311
Query: 467 TSAFEEIVSKENDVSKEDAVRWIRALEKCGKYHIEAW 503
A I+ ++ + A ++ L+ G+Y + W
Sbjct: 1312HRALHTILQEQGSLDSSKAESMVKNLQTTGRYLRDVW 1422
>TC216793 similar to UP|FENS_PEA (Q41014) Ferredoxin--NADP reductase, root
isozyme, chloroplast precursor (FNR) , partial (97%)
Length = 1408
Score = 65.9 bits (159), Expect = 4e-11
Identities = 55/200 (27%), Positives = 90/200 (44%), Gaps = 9/200 (4%)
Frame = +3
Query: 313 KKGLCSSWLAALDPRDAVSLPVWFQKGSL-PTPSPSLPLILVGPGTGCAPFRGFIEERAL 371
K G+CS++L P D + + K L P P+ I++ GTG APFRG++ +
Sbjct: 777 KNGICSNFLCNSKPGDKIQITGPSGKIMLLPEDDPNATHIMIATGTGVAPFRGYLRRMFM 956
Query: 372 QS-KTISIAPIIF-FFGCWNEDGDFLYKDF--WLNHSQNNGVLSESTGGGFYVAFSRDQP 427
+S T + + F G N D ++F +LN +N A SR+Q
Sbjct: 957 ESVPTYKFGGLAWLFLGVANTDSLLYDEEFSKYLNDYSDNFRYDR--------ALSREQK 1112
Query: 428 E----KVYVQHKLREHSGRVWNLLAEGASVYIAGSLTKMPTDVTSAFEEIVSKENDVSKE 483
K+YVQ K+ E+S ++ LL GA +Y G MP ++ + K + E
Sbjct: 1113NKNGGKMYVQDKIEEYSDEIFKLLDNGAHIYFCGLKGMMP-----GIQDTLKKVAEQRGE 1277
Query: 484 DAVRWIRALEKCGKYHIEAW 503
+ L+K ++H+E +
Sbjct: 1278SWEEKLSQLKKNKQWHVEVY 1337
>TC215293 similar to UP|FENR_TOBAC (O04977) Ferredoxin--NADP reductase,
leaf-type isozyme, chloroplast precursor (FNR) ,
complete
Length = 1488
Score = 59.3 bits (142), Expect = 4e-09
Identities = 59/236 (25%), Positives = 105/236 (44%), Gaps = 11/236 (4%)
Frame = +1
Query: 279 KKREFSISSS---QSSHPNQVHLTVSVVSWTTPYKRKKKGLCSSWLAALDPRDAVSLPVW 335
K R +SI+SS V L V + +T KG+CS++L L P V++
Sbjct: 553 KLRLYSIASSAIGDFGDSKTVSLCVKRLVYTNENGEIVKGVCSNFLCDLKPGAEVTITGP 732
Query: 336 FQKGSLPTPSPSLPLILVGPGTGCAPFRGFIEERALQS-KTISIAPIIFFFGCWNEDGDF 394
K L P+ +I++G GTG APFR F+ + + + + + F
Sbjct: 733 VGKEMLMPKDPNATIIMLGTGTGIAPFRSFLWKMFFEKHEDYKFNGLAWLFLGVPTSSSL 912
Query: 395 LYKDFWLNHSQNNGVLSESTGGGFYVAF--SRDQP----EKVYVQHKLREHSGRVWNLL- 447
LYK+ + + E + F + F SR+Q EK+Y+Q ++ +++ +W LL
Sbjct: 913 LYKEEFEK-------MQEKSPENFRLDFAVSREQTNEKGEKMYIQTRMAQYAEELWELLK 1071
Query: 448 AEGASVYIAGSLTKMPTDVTSAFEEIVSKENDVSKEDAVRWIRALEKCGKYHIEAW 503
+ VY+ G L M + + +K+ D + R L+K ++++E +
Sbjct: 1072KDNTFVYMCG-LKGMEKGIDDIMVSLAAKDG----IDWTEYKRQLKKAEQWNVEVY 1224
>TC215292 similar to UP|FENR_PEA (P10933) Ferredoxin--NADP reductase, leaf
isozyme, chloroplast precursor (FNR) , complete
Length = 1435
Score = 57.4 bits (137), Expect = 1e-08
Identities = 60/239 (25%), Positives = 102/239 (42%), Gaps = 14/239 (5%)
Frame = +2
Query: 279 KKREFSISSS---QSSHPNQVHLTVSVVSWTTPYKRKKKGLCSSWLAALDPRDAVSLPVW 335
K R +SI+SS V L V + +T KG+CS++L L P V +
Sbjct: 545 KLRLYSIASSALGDFGDSKTVSLCVKRLVYTNENGELVKGVCSNFLCDLKPGAEVKITGP 724
Query: 336 FQKGSLPTPSPSLPLILVGPGTGCAPFRGFIEERALQS-KTISIAPIIFFFGCWNEDGDF 394
K L P+ +I++ GTG APFR F+ + + + + F
Sbjct: 725 VGKEMLMPKDPNATVIMLATGTGIAPFRSFLWKMFFEKHDDYKFNGLAWLFLGVPTSSSL 904
Query: 395 LYKDFWLNHSQNNGVLSESTGGGFYVAF--SRDQP----EKVYVQHKLREHSGRVWNLL- 447
LYK+ + + E F + F SR+Q EK+Y+Q ++ +++ +W LL
Sbjct: 905 LYKEEFEK-------MKEKAPDNFRLDFAVSREQTNEKGEKMYIQTRMAQYAEELWELLK 1063
Query: 448 AEGASVYIAGSLTKMPTDVTSAFEEIVSKENDVSKEDAVRWI---RALEKCGKYHIEAW 503
+ VY+ G L M + + +K D + WI R L+K ++++E +
Sbjct: 1064KDNTFVYMCG-LKGMEKGIDDIMVSLAAK-------DGIDWIEYKRQLKKAEQWNVEVY 1216
>TC225902 similar to UP|O48937 (O48937) NADPH cytochrome P450 reductase ,
partial (11%)
Length = 581
Score = 50.8 bits (120), Expect = 1e-06
Identities = 20/73 (27%), Positives = 38/73 (51%)
Frame = +2
Query: 431 YVQHKLREHSGRVWNLLAEGASVYIAGSLTKMPTDVTSAFEEIVSKENDVSKEDAVRWIR 490
YVQHK+ E + +W++++ GA +Y+ G M +V A I+ ++ + A +
Sbjct: 2 YVQHKMMEKASEIWSMISXGAYIYVCGDAKGMARNVHRALHTILQEQGSLDSSKAESMVN 181
Query: 491 ALEKCGKYHIEAW 503
L+ G+Y + W
Sbjct: 182 NLQTTGRYLRDVW 220
>AW350962 homologue to GP|27261128|gb| NADPH:P450 reductase {Glycine max},
partial (10%)
Length = 519
Score = 47.0 bits (110), Expect = 2e-05
Identities = 19/66 (28%), Positives = 35/66 (52%)
Frame = -2
Query: 433 QHKLREHSGRVWNLLAEGASVYIAGSLTKMPTDVTSAFEEIVSKENDVSKEDAVRWIRAL 492
QHK+ + + +WNL+++G +Y+ G M DV IV ++ +V A ++ L
Sbjct: 518 QHKMMDKAANLWNLISQGGYLYVCGDAKGMAXDVXRTLHTIVQQQENVDSSKAEGIVKKL 339
Query: 493 EKCGKY 498
+ G+Y
Sbjct: 338 QMDGRY 321
>BE347814 similar to SP|P41346|FENR Ferredoxin--NADP reductase chloroplast
precursor (EC 1.18.1.2) (FNR). [Broad bean] {Vicia
faba}, partial (51%)
Length = 613
Score = 45.1 bits (105), Expect = 8e-05
Identities = 29/89 (32%), Positives = 44/89 (48%), Gaps = 3/89 (3%)
Frame = +2
Query: 281 REFSISSS---QSSHPNQVHLTVSVVSWTTPYKRKKKGLCSSWLAALDPRDAVSLPVWFQ 337
R +SI+SS V L V + +T G+CS++L L P V++ V
Sbjct: 74 RLYSIASSAIGDFGDSKTVSLCVKRLGFTNENGEMVYGVCSNFLCDLKPAAEVTITVPVG 253
Query: 338 KGSLPTPSPSLPLILVGPGTGCAPFRGFI 366
K L P+ +I++G GTG AP+R F+
Sbjct: 254 KEMLMPKDPNATIIMLGTGTGIAPYRSFL 340
>TC216794 homologue to UP|FENS_PEA (Q41014) Ferredoxin--NADP reductase, root
isozyme, chloroplast precursor (FNR) , partial (37%)
Length = 422
Score = 40.0 bits (92), Expect = 0.002
Identities = 21/62 (33%), Positives = 33/62 (52%), Gaps = 1/62 (1%)
Frame = +2
Query: 313 KKGLCSSWLAALDPRDAVSLPVWFQKGSL-PTPSPSLPLILVGPGTGCAPFRGFIEERAL 371
K G+CS++L P D + + K L P P+ I++ GTG APFRG++ +
Sbjct: 59 KNGICSNFLCNSKPGDKIQITGPSGKIMLLPEDDPNATHIMIATGTGVAPFRGYLRRMFM 238
Query: 372 QS 373
+S
Sbjct: 239 ES 244
>TC217618 similar to GB|AAO64868.1|29028978|BT005933 At5g50860 {Arabidopsis
thaliana;} , partial (60%)
Length = 1325
Score = 32.0 bits (71), Expect = 0.67
Identities = 16/35 (45%), Positives = 22/35 (62%), Gaps = 1/35 (2%)
Frame = +1
Query: 243 LYQYNQKERRTVLEVLKDFPSVQMPL-EWLIQLVP 276
LY+ Q +R +LE KDFPS +PL E L+ + P
Sbjct: 538 LYKPQQPYKRNILETFKDFPSSSLPLIETLLAIDP 642
>TC226542 weakly similar to UP|Q9ZWQ5 (Q9ZWQ5) UDP-glycose:flavonoid
glycosyltransferase, partial (18%)
Length = 753
Score = 29.6 bits (65), Expect = 3.3
Identities = 18/68 (26%), Positives = 33/68 (48%), Gaps = 2/68 (2%)
Frame = +1
Query: 392 GDFLYKDFWLNHSQNNGVLSESTGGGFYVAFSRDQPE--KVYVQHKLREHSGRVWNLLAE 449
G+ YK +W+ Q+ G+ S + G + SRD+ + ++ + L S W+ L
Sbjct: 379 GEECYKGWWIFAQQHEGIDSGAQGDQAFQGGSRDRTQSLRLVLLFHLLNKSNVFWDTLIS 558
Query: 450 GASVYIAG 457
+S+ I G
Sbjct: 559 LSSISIPG 582
>BM731788
Length = 423
Score = 29.6 bits (65), Expect = 3.3
Identities = 21/64 (32%), Positives = 31/64 (47%), Gaps = 4/64 (6%)
Frame = +3
Query: 342 PTPSPSLPLILVGPGTGCAPFRGFIEERALQSK----TISIAPIIFFFGCWNEDGDFLYK 397
P P+PSL L+ P P R F+E Q + T + + FFF + + G FL+
Sbjct: 114 PNPNPSLRG-LISP-----PIRVFLEPNLKQKRVSFNTKAKTVVFFFFFYFRDLGSFLFI 275
Query: 398 DFWL 401
FW+
Sbjct: 276 YFWI 287
>TC223433 weakly similar to UP|Q9AST4 (Q9AST4) AT4g31930/F10N7_260, partial
(36%)
Length = 414
Score = 28.9 bits (63), Expect = 5.6
Identities = 18/64 (28%), Positives = 26/64 (40%), Gaps = 5/64 (7%)
Frame = -3
Query: 346 PSLPLILVGPGTGCAPFRGFIEERALQSKTISIAPIIFFFGCWNEDGDF-----LYKDFW 400
P P + + P T C + +EER L F CWN+ + L + FW
Sbjct: 295 PLAPHVALQP*TNCIESQAGVEERILLDTRFRFGE---FV*CWNQRAIWSRVLVLLRRFW 125
Query: 401 LNHS 404
L H+
Sbjct: 124 LRHA 113
>CO984402
Length = 737
Score = 28.5 bits (62), Expect = 7.4
Identities = 15/50 (30%), Positives = 28/50 (56%), Gaps = 2/50 (4%)
Frame = -1
Query: 34 YEGTLDPWLSSLWRMLNMIKPELLPSGPDVLIQDTTL--IDQPKVQITYH 81
Y+ TLD LSS++ ++ P+ + DV ++D T+ +++ VQ H
Sbjct: 686 YDVTLDVCLSSVFSQTKVLNPQQVTETIDVCVEDETVNYLNRKDVQSAMH 537
>AW830539 similar to SP|O04450|TCPE_ T-complex protein 1 epsilon subunit
(TCP-1-epsilon) (CCT-epsilon). [Mouse-ear cress],
partial (13%)
Length = 238
Score = 28.5 bits (62), Expect = 7.4
Identities = 13/37 (35%), Positives = 19/37 (51%)
Frame = +1
Query: 383 FFFGCWNEDGDFLYKDFWLNHSQNNGVLSESTGGGFY 419
F+ G W G Y +W N S + V+S + G G+Y
Sbjct: 106 FYMGWWRRIGIDSYTYWWNNCSYVSRVISTNLGNGWY 216
>NP164640 receptor-like protein kinase
Length = 828
Score = 28.1 bits (61), Expect = 9.6
Identities = 17/56 (30%), Positives = 27/56 (47%), Gaps = 1/56 (1%)
Frame = +2
Query: 289 QSSHPNQVHL-TVSVVSWTTPYKRKKKGLCSSWLAALDPRDAVSLPVWFQKGSLPT 343
QSSH Q + TVS + P++ K + L S+W + +S + Q S P+
Sbjct: 155 QSSHSTQNSVATVSPSPFHAPHRLKMRTLASTWASLTQTMWGISQTTYLQWNSTPS 322
>TC217080 similar to UP|Q94EJ2 (Q94EJ2) At1g08460/T27G7_7 (HDA8), partial
(88%)
Length = 1303
Score = 28.1 bits (61), Expect = 9.6
Identities = 24/85 (28%), Positives = 37/85 (43%)
Frame = +3
Query: 87 VSHSSDTGSVRSMHPGKSSSNRNGYPDCFLKLVKNLPLTKPNSGKDVRHFEFEFVSHAIE 146
+S + GS HP S + G + + N+PL K H E V +I+
Sbjct: 651 ISLHMNHGSWGPSHPQSGSVDELGEGEGY-GFNLNIPLPNGTGDKGYVHAFNELVVPSIQ 827
Query: 147 YDTGDILEVLPGQDSAAVDAFIRRC 171
D++ ++ GQDS A D R+C
Sbjct: 828 KFGPDMIVLVLGQDSNAFDPNGRQC 902
>TC218901 similar to UP|Q9LGX2 (Q9LGX2) ESTs AU082689(E31988), partial (56%)
Length = 1058
Score = 28.1 bits (61), Expect = 9.6
Identities = 12/34 (35%), Positives = 22/34 (64%)
Frame = -3
Query: 279 KKREFSISSSQSSHPNQVHLTVSVVSWTTPYKRK 312
K+REF+ ++ H +VH+++ S+TT Y+ K
Sbjct: 777 KQREFAS*ITKQIHDQKVHISLQETSFTTNYQPK 676
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.136 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,020,654
Number of Sequences: 63676
Number of extensions: 381802
Number of successful extensions: 1913
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 1886
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1894
length of query: 504
length of database: 12,639,632
effective HSP length: 101
effective length of query: 403
effective length of database: 6,208,356
effective search space: 2501967468
effective search space used: 2501967468
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148995.5


