

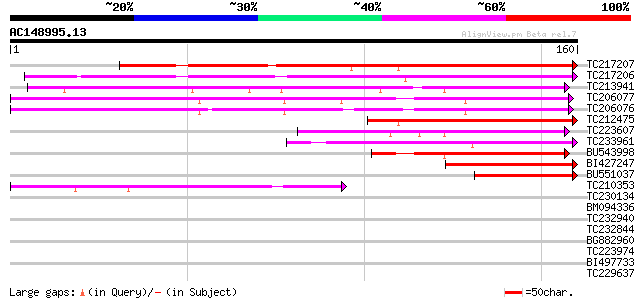
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148995.13 - phase: 0
(160 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC217207 119 6e-28
TC217206 similar to GB|AAM16233.1|20334744|AY093972 AT3g45210/T1... 110 2e-25
TC213941 similar to GB|AAO39924.1|28372884|BT003696 At4g04630 {A... 95 2e-20
TC206077 similar to UP|O80698 (O80698) F8K4.12, partial (31%) 86 8e-18
TC206076 similar to UP|O80698 (O80698) F8K4.12, partial (43%) 85 2e-17
TC212475 weakly similar to GB|AAM16233.1|20334744|AY093972 AT3g4... 79 1e-15
TC223607 similar to GB|AAO39924.1|28372884|BT003696 At4g04630 {A... 70 6e-13
TC233961 similar to UP|Q944R0 (Q944R0) AT3g15040/K15M2_18, parti... 67 4e-12
BU543998 60 4e-10
BI427247 57 3e-09
BU551037 57 3e-09
TC210353 50 5e-07
TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Pro... 37 0.005
BM094336 32 0.17
TC232940 homologue to UP|Q7XVJ5 (Q7XVJ5) OJ000126_13.10 protein,... 31 0.29
TC232844 similar to UP|Q9SP09 (Q9SP09) DnaJ-like protein, partia... 28 1.9
BG882960 28 2.4
TC223974 28 2.4
BI497733 28 2.4
TC229637 similar to UP|KIWI_ARATH (O65154) RNA polymerase II tra... 27 3.2
>TC217207
Length = 737
Score = 119 bits (298), Expect = 6e-28
Identities = 67/133 (50%), Positives = 90/133 (67%), Gaps = 4/133 (3%)
Frame = +2
Query: 32 EFDEADVWNMSYSNSNTNIEPKKGVPGLKRVSRKMEANNKVNPLASSSLPMNIPDWSKIL 91
EF E+D++N + +NS E K V + + + +V A +SLP+N+PDWSKIL
Sbjct: 131 EFHESDLYNSARANSP---ELAKSVRSSRFHNYSSSSAARVG--APASLPVNVPDWSKIL 295
Query: 92 KEEY-KKKKESSDDEDEG---DYDGVVQLPPHEYLARTRGASLSVHEGKGRTLKGRDLRS 147
+EY + ++ + DD+DE + DGV ++PPHE+LARTR AS SVHEG GRTLKGRDL
Sbjct: 296 GDEYGRNQRRNYDDDDEARSDEEDGVGRVPPHEFLARTRIASFSVHEGVGRTLKGRDLSR 475
Query: 148 VRNAIWKKVGFED 160
VRNAIW K GF+D
Sbjct: 476 VRNAIWAKTGFQD 514
>TC217206 similar to GB|AAM16233.1|20334744|AY093972 AT3g45210/T14D3_150
{Arabidopsis thaliana;} , partial (44%)
Length = 721
Score = 110 bits (276), Expect = 2e-25
Identities = 66/157 (42%), Positives = 92/157 (58%), Gaps = 1/157 (0%)
Frame = +2
Query: 5 RKSFLSRTSYIFPETNFNQKSSQGKELEFDEADVWNMSYSNSNTNIEPKKGVPGLKRVSR 64
R + + R +Y F + S EF E+D++N + +NS E +K V + +
Sbjct: 59 RMTNIRRATYRFLPA-MDTDSFSDSNFEFQESDLYNSARANSP---EFRKSVRASRFHNY 226
Query: 65 KMEANNKVNPLASSSLPMNIPDWSKILKEEYKKKKESSDDEDEGDY-DGVVQLPPHEYLA 123
P+ SLP+N+PDWSKIL +E+ + + + DE + D DG ++PPHE+LA
Sbjct: 227 SSSGGRVGTPV---SLPVNVPDWSKILGDEFGRNQRRNYDEAQSDEEDGDGRVPPHEFLA 397
Query: 124 RTRGASLSVHEGKGRTLKGRDLRSVRNAIWKKVGFED 160
+T AS SVHEG GRTLKGRDL VRNAIW K GF+D
Sbjct: 398 KTGIASFSVHEGVGRTLKGRDLSRVRNAIWAKTGFQD 508
>TC213941 similar to GB|AAO39924.1|28372884|BT003696 At4g04630 {Arabidopsis
thaliana;} , partial (40%)
Length = 776
Score = 94.7 bits (234), Expect = 2e-20
Identities = 70/187 (37%), Positives = 93/187 (49%), Gaps = 34/187 (18%)
Frame = +2
Query: 6 KSFLSRTSY--IFPETNFNQKSSQGKELEFDEADVWNMSYSNSNTNI----------EPK 53
K ++ T Y +F ++ + K+ +FDE DVW+++ +S
Sbjct: 83 KGLITFTIYRGVFLNKSWGVGGREMKDRDFDEEDVWSVANDDSGKETTRACKESCGSSSS 262
Query: 54 KGVPGLKRVSRKM-EANNKVNPL----------ASSSLPMNIPDWSKILKEEYKKKKESS 102
L RK+ ANN NPL SSS PM+IPDWSKI + KK +
Sbjct: 263 SSAWRLPAAPRKIPRANNNANPLPAASDAPLVKGSSSAPMDIPDWSKIYGKSCKKGSTAD 442
Query: 103 D-------DEDEGDYDGVVQLPPHEY----LARTRGASLSVHEGKGRTLKGRDLRSVRNA 151
D D+D+ D D +V PPHE+ LAR++ +S SV EG GRTLKGRDL VRNA
Sbjct: 443 DGASNKGGDDDDDDDDDMV--PPHEWIARKLARSQISSFSVCEGMGRTLKGRDLSKVRNA 616
Query: 152 IWKKVGF 158
I K GF
Sbjct: 617 ILTKTGF 637
>TC206077 similar to UP|O80698 (O80698) F8K4.12, partial (31%)
Length = 780
Score = 85.9 bits (211), Expect = 8e-18
Identities = 61/199 (30%), Positives = 88/199 (43%), Gaps = 40/199 (20%)
Frame = +3
Query: 1 MASSRKSFLSRTSYIFPETNFNQKSSQGKELEFDEADVWNMSYSNSNTNIEP-----KKG 55
MA RK SR+ ++Q S+ E E DVW +P G
Sbjct: 96 MAKGRKLTTSRSERFLGTYAYSQGSAAVNPSELREEDVWGAGDDAGEREWDPHFAAMSNG 275
Query: 56 VPGLKRVSRKMEANNKVNPLA---------------------------------SSSLPM 82
+R+ R + + +V L+ ++S PM
Sbjct: 276 GGSRRRIPRDTDVHRRVGGLSLAFEAPASGASPRIVHQFRAREEMASTPRVRHMATSAPM 455
Query: 83 NIPDWSKILK-EEYKKKKESSDDEDEGDYDGVVQLPPHEYLARTRG-ASLSVHEGKGRTL 140
N+PDWSKIL+ + + +DEDE + +PPHEYLAR++ + SV EG GRTL
Sbjct: 456 NVPDWSKILRVDSVDSLNDDYNDEDESE-----MVPPHEYLARSQTMVANSVFEGVGRTL 620
Query: 141 KGRDLRSVRNAIWKKVGFE 159
KGRDL VR+A+W + GF+
Sbjct: 621 KGRDLSRVRDAVWSQTGFD 677
>TC206076 similar to UP|O80698 (O80698) F8K4.12, partial (43%)
Length = 1225
Score = 84.7 bits (208), Expect = 2e-17
Identities = 64/199 (32%), Positives = 91/199 (45%), Gaps = 40/199 (20%)
Frame = +3
Query: 1 MASSRKSFLSRTSYIFPETNFNQKSSQGKELEFDEADVWNMSYSNSNTNIEP------KK 54
MA RK SR+ ++Q S+ E E DVW + EP
Sbjct: 135 MAKGRKLTTSRSERFLGTYAYSQGSAALNPSELREEDVWGAGDDAGDREWEPHAAALNNS 314
Query: 55 GVPGLKRVSRKMEANNKVNPLA---------------------------------SSSLP 81
GV +R+ R E + +V L+ ++S P
Sbjct: 315 GV-SRRRIPRDTEEHRRVGGLSLAFEAPASGSSPRMVHQFRAREEMTSTPRGRHMATSAP 491
Query: 82 MNIPDWSKILKEEYKKKKESSDDEDEGDYDGVVQLPPHEYLARTRG-ASLSVHEGKGRTL 140
+N+PDWSKIL+ + ES +DED + + +PPHEYLAR++ + SV EG GRTL
Sbjct: 492 VNVPDWSKILRVD---SVESINDEDGDESE---MMPPHEYLARSQKMVANSVFEGVGRTL 653
Query: 141 KGRDLRSVRNAIWKKVGFE 159
KGRDL VR+A+W + GF+
Sbjct: 654 KGRDLSRVRDAVWNQTGFD 710
>TC212475 weakly similar to GB|AAM16233.1|20334744|AY093972
AT3g45210/T14D3_150 {Arabidopsis thaliana;} , partial
(35%)
Length = 458
Score = 78.6 bits (192), Expect = 1e-15
Identities = 38/60 (63%), Positives = 47/60 (78%), Gaps = 1/60 (1%)
Frame = +3
Query: 102 SDDEDEG-DYDGVVQLPPHEYLARTRGASLSVHEGKGRTLKGRDLRSVRNAIWKKVGFED 160
+DD +EG D + ++PPHE+LAR R AS SVHEG GRTLKGRDL ++RNAIW K GF+D
Sbjct: 6 NDDNNEGYDDERSGRVPPHEFLARNRVASFSVHEGVGRTLKGRDLSTLRNAIWAKTGFQD 185
>TC223607 similar to GB|AAO39924.1|28372884|BT003696 At4g04630 {Arabidopsis
thaliana;} , partial (29%)
Length = 617
Score = 69.7 bits (169), Expect = 6e-13
Identities = 44/98 (44%), Positives = 54/98 (54%), Gaps = 21/98 (21%)
Frame = +3
Query: 82 MNIPDWSKILKEEYKKKKESSDDED-----------EGDYDGVV------QLPPHEY--- 121
+NIPDWSKI + + K S D+D + D DG +LPPHE+
Sbjct: 15 VNIPDWSKIYRTKTPKNSVSRFDDDYDGDGAANYGGDSDEDGEENDESDSKLPPHEFIAR 194
Query: 122 -LARTRGASLSVHEGKGRTLKGRDLRSVRNAIWKKVGF 158
LAR+R +S SV EG GRTLKGRDL VRN + K GF
Sbjct: 195 RLARSRISSFSVLEGAGRTLKGRDLSKVRNDVLSKTGF 308
>TC233961 similar to UP|Q944R0 (Q944R0) AT3g15040/K15M2_18, partial (25%)
Length = 487
Score = 67.0 bits (162), Expect = 4e-12
Identities = 38/86 (44%), Positives = 48/86 (55%), Gaps = 4/86 (4%)
Frame = +3
Query: 79 SLPMNIPDWSKILKEEYKKKKESSDDEDEGDYDGVVQLPPHEYLARTRGAS----LSVHE 134
S P+N+P ++ +E DD+ D LPPHE +AR S SV E
Sbjct: 219 SAPVNVP----LMPMRRHHHREFDDDDGNEDAAEEEMLPPHEIVARNSAQSPMLAYSVLE 386
Query: 135 GKGRTLKGRDLRSVRNAIWKKVGFED 160
G GRTLKGRDLR VRNA+W++ GF D
Sbjct: 387 GVGRTLKGRDLRXVRNAVWRQTGFLD 464
>BU543998
Length = 571
Score = 60.1 bits (144), Expect = 4e-10
Identities = 35/60 (58%), Positives = 40/60 (66%), Gaps = 4/60 (6%)
Frame = -1
Query: 103 DDEDEGDYDGVVQLPPHEY----LARTRGASLSVHEGKGRTLKGRDLRSVRNAIWKKVGF 158
DDEDE D +PPHE+ LAR++ +S SV EG GRTLKGRDL VRNAI K GF
Sbjct: 553 DDEDEDD-----MIPPHEWIARKLARSQISSFSVCEGIGRTLKGRDLSKVRNAILTKTGF 389
>BI427247
Length = 316
Score = 57.4 bits (137), Expect = 3e-09
Identities = 27/37 (72%), Positives = 29/37 (77%)
Frame = +3
Query: 124 RTRGASLSVHEGKGRTLKGRDLRSVRNAIWKKVGFED 160
RT+ S SVHEG GRTLKGRDL VRNAIW K GF+D
Sbjct: 18 RTKRFSSSVHEGVGRTLKGRDLSRVRNAIWAKTGFQD 128
>BU551037
Length = 173
Score = 57.4 bits (137), Expect = 3e-09
Identities = 26/29 (89%), Positives = 27/29 (92%)
Frame = -2
Query: 132 VHEGKGRTLKGRDLRSVRNAIWKKVGFED 160
+HEG G TLKGRDLRSVRNAIWKKVGFED
Sbjct: 172 MHEGIGSTLKGRDLRSVRNAIWKKVGFED 86
>TC210353
Length = 438
Score = 50.1 bits (118), Expect = 5e-07
Identities = 31/100 (31%), Positives = 55/100 (55%), Gaps = 5/100 (5%)
Frame = +3
Query: 1 MASSRKSFLSRTSYIFP-ETNFNQKSSQGKELE----FDEADVWNMSYSNSNTNIEPKKG 55
MAS + F SR+ P ++ + S +LE FDE++++N + +N++
Sbjct: 147 MASRKNHFTSRSHRFLPVASDIHDSVSLTMDLESAFEFDESEIYNSARANNSFEFRRSLH 326
Query: 56 VPGLKRVSRKMEANNKVNPLASSSLPMNIPDWSKILKEEY 95
G ++K +++ +P +S+P+NIPDWSKIL +EY
Sbjct: 327 GRGSNSSAKKKPSSSSGDP---ASVPVNIPDWSKILGDEY 437
>TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23),
partial (8%)
Length = 581
Score = 36.6 bits (83), Expect = 0.005
Identities = 23/86 (26%), Positives = 42/86 (48%), Gaps = 5/86 (5%)
Frame = +2
Query: 29 KELEFDEADVWNMSYSNS---NTNIEPKKGVPGLKRVSRKM--EANNKVNPLASSSLPMN 83
+ L F+E +W S+ + +T + K + V+ + +ANN V+P+ P++
Sbjct: 47 QSLSFEENTLWVGSFVEALVLDTLLATAKSLACFLVVTGSLLTDANNLVSPMVDGRFPLD 226
Query: 84 IPDWSKILKEEYKKKKESSDDEDEGD 109
P K EE K ++ DD+D+ D
Sbjct: 227 QPCKGKHTSEENKDASDTEDDDDDDD 304
>BM094336
Length = 284
Score = 31.6 bits (70), Expect = 0.17
Identities = 15/35 (42%), Positives = 20/35 (56%)
Frame = +1
Query: 79 SLPMNIPDWSKILKEEYKKKKESSDDEDEGDYDGV 113
S P+NIPDWS+I + + K S GDYD +
Sbjct: 61 SAPLNIPDWSQIYRNKSNKTTPKSVSR-FGDYDDI 162
Score = 26.2 bits (56), Expect = 7.1
Identities = 10/20 (50%), Positives = 15/20 (75%)
Frame = +3
Query: 102 SDDEDEGDYDGVVQLPPHEY 121
SDDE+E + + +LPPHE+
Sbjct: 225 SDDEEEEEDEYDTKLPPHEF 284
>TC232940 homologue to UP|Q7XVJ5 (Q7XVJ5) OJ000126_13.10 protein, partial
(13%)
Length = 796
Score = 30.8 bits (68), Expect = 0.29
Identities = 15/24 (62%), Positives = 16/24 (66%)
Frame = +3
Query: 132 VHEGKGRTLKGRDLRSVRNAIWKK 155
V EG GRTLKGRDLR V+ K
Sbjct: 645 VLEGVGRTLKGRDLRQVKKPFCAK 716
>TC232844 similar to UP|Q9SP09 (Q9SP09) DnaJ-like protein, partial (35%)
Length = 466
Score = 28.1 bits (61), Expect = 1.9
Identities = 19/73 (26%), Positives = 34/73 (46%), Gaps = 2/73 (2%)
Frame = +2
Query: 54 KGVPGLKRVSRKM-EANNKVNPLASSSLPMNI-PDWSKILKEEYKKKKESSDDEDEGDYD 111
KG ++SR++ +A+ K+ P+ L +N+ P W K K + +K + D
Sbjct: 137 KGTTKKMKISREIADASGKIMPV-EEILTINVKPGWKKGTKITFPEKGNEQPNVTPADLV 313
Query: 112 GVVQLPPHEYLAR 124
++ PH AR
Sbjct: 314 FIIDEKPHSVFAR 352
>BG882960
Length = 386
Score = 27.7 bits (60), Expect = 2.4
Identities = 13/28 (46%), Positives = 20/28 (71%), Gaps = 1/28 (3%)
Frame = +3
Query: 131 SVHEGKGRTLKGRDLRSV-RNAIWKKVG 157
S+HEGKG+T G+D R ++++ KK G
Sbjct: 147 SIHEGKGKTPGGKDSRDFQQSSVCKKGG 230
>TC223974
Length = 429
Score = 27.7 bits (60), Expect = 2.4
Identities = 14/35 (40%), Positives = 19/35 (54%)
Frame = -3
Query: 95 YKKKKESSDDEDEGDYDGVVQLPPHEYLARTRGAS 129
++ + + E EGD DGVVQ P +AR R S
Sbjct: 421 FENGEVEGEGEGEGDLDGVVQR*PDVGVARRRSGS 317
>BI497733
Length = 427
Score = 27.7 bits (60), Expect = 2.4
Identities = 11/19 (57%), Positives = 15/19 (78%)
Frame = -1
Query: 42 SYSNSNTNIEPKKGVPGLK 60
SYSN+N+N+ KKG+ LK
Sbjct: 370 SYSNANSNLSRKKGLQSLK 314
>TC229637 similar to UP|KIWI_ARATH (O65154) RNA polymerase II transcriptional
coactivator KIWI, partial (55%)
Length = 743
Score = 27.3 bits (59), Expect = 3.2
Identities = 19/62 (30%), Positives = 29/62 (46%), Gaps = 1/62 (1%)
Frame = +1
Query: 64 RKMEANNKVNPLASSSLPMNIPDWSKI-LKEEYKKKKESSDDEDEGDYDGVVQLPPHEYL 122
+K+ A +NPL SS + + K L E+ K + DD+D D D PP + L
Sbjct: 106 KKLFARVFLNPLTHSSHNSHTTETEKKNLLEKMSGKAKRRDDDDASDADSEGHAPPMKSL 285
Query: 123 AR 124
+
Sbjct: 286 KK 291
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.308 0.128 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,643,798
Number of Sequences: 63676
Number of extensions: 78765
Number of successful extensions: 489
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 466
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 477
length of query: 160
length of database: 12,639,632
effective HSP length: 90
effective length of query: 70
effective length of database: 6,908,792
effective search space: 483615440
effective search space used: 483615440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 55 (25.8 bits)
Medicago: description of AC148995.13


