

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148995.1 + phase: 0
(220 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
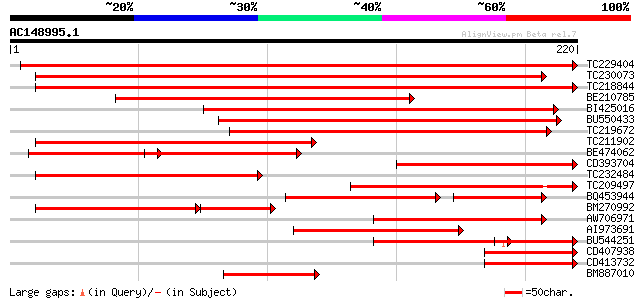
Score E
Sequences producing significant alignments: (bits) Value
TC229404 weakly similar to GB|AAS56573.1|45270384|AY558247 YJL05... 410 e-115
TC230073 similar to UP|Q9ASW6 (Q9ASW6) T3B23.2/T3B23.2, partial ... 348 1e-96
TC218844 weakly similar to GB|AAS56573.1|45270384|AY558247 YJL05... 329 7e-91
BE210785 229 1e-60
BI425016 similar to GP|20197602|gb expressed protein {Arabidopsi... 202 1e-52
BU550433 similar to GP|28372900|gb At5g06300 {Arabidopsis thalia... 202 1e-52
TC219672 similar to UP|Q9ASW6 (Q9ASW6) T3B23.2/T3B23.2, partial ... 200 4e-52
TC211902 similar to UP|Q84MC2 (Q84MC2) At5g11950, partial (51%) 168 2e-42
BE474062 similar to GP|9758410|dbj lysine decarboxylase-like pro... 94 3e-35
CD393704 139 8e-34
TC232484 similar to UP|Q9FNH8 (Q9FNH8) Lysine decarboxylase-like... 138 2e-33
TC209497 similar to UP|Q9ASW6 (Q9ASW6) T3B23.2/T3B23.2, partial ... 136 9e-33
BQ453944 similar to GP|20804658|dbj lysine decarboxylase-like pr... 103 4e-29
BM270992 similar to GP|9758410|dbj| lysine decarboxylase-like pr... 99 7e-29
AW706971 similar to GP|13605607|gb| T3B23.2/T3B23.2 {Arabidopsis... 110 4e-25
AI973691 similar to GP|28372900|gb| At5g06300 {Arabidopsis thali... 109 1e-24
BU544251 94 5e-20
CD407938 homologue to PIR|S04125|S041 chlorophyll a/b-binding pr... 74 4e-14
CD413732 homologue to PIR|E96640|E966 PSI type III chlorophyll a... 74 4e-14
BM887010 69 2e-12
>TC229404 weakly similar to GB|AAS56573.1|45270384|AY558247 YJL055W
{Saccharomyces cerevisiae;} , partial (32%)
Length = 1124
Score = 410 bits (1054), Expect = e-115
Identities = 200/216 (92%), Positives = 211/216 (97%)
Frame = +2
Query: 5 NGEIKLSKFKRICVFCGSSPGKKTTYQDAAMNLGNELVSRNIDLVYGGGSIGLMGLVSQA 64
+ EI++SKFKR+CVFCGSSPGKK +YQDAA+ LGNELVSRNIDLVYGGGSIGLMGLVSQA
Sbjct: 251 HAEIRVSKFKRVCVFCGSSPGKKRSYQDAAIELGNELVSRNIDLVYGGGSIGLMGLVSQA 430
Query: 65 VHDGGRHVIGVIPKTLMPRELTGETVGEVKAVADMHQRKAEMAKHSDAFIALPGGYGTLE 124
VHDGGRHVIGVIPKTLMPRELTGETVGEVKAVADMHQRKAEMAKHSDAFIALPGGYGTLE
Sbjct: 431 VHDGGRHVIGVIPKTLMPRELTGETVGEVKAVADMHQRKAEMAKHSDAFIALPGGYGTLE 610
Query: 125 ELLEVITWAQLGIHDKPVGLVNVDGYFNSLLSFIDKAVEEGFISPNARHIIVSAPTAKEL 184
ELLEVITWAQLGIHDKPVGLVNVDGYFNSLLSFIDKAVEEGFISPNARHIIVSAPT+KEL
Sbjct: 611 ELLEVITWAQLGIHDKPVGLVNVDGYFNSLLSFIDKAVEEGFISPNARHIIVSAPTSKEL 790
Query: 185 VKKLEEYVPCHEGVASKLSWQMEQQLAYPQDYDISR 220
VKKLE+YVPCHE VASKLSWQ+EQQL YP++YDISR
Sbjct: 791 VKKLEDYVPCHESVASKLSWQIEQQLTYPEEYDISR 898
>TC230073 similar to UP|Q9ASW6 (Q9ASW6) T3B23.2/T3B23.2, partial (93%)
Length = 863
Score = 348 bits (893), Expect = 1e-96
Identities = 171/198 (86%), Positives = 183/198 (92%)
Frame = +2
Query: 11 SKFKRICVFCGSSPGKKTTYQDAAMNLGNELVSRNIDLVYGGGSIGLMGLVSQAVHDGGR 70
SKFKRICVFCGSSPG KT+Y DAA+ LG ELVSRNIDLVYGGGSIGLMGL+S AV++GGR
Sbjct: 2 SKFKRICVFCGSSPGNKTSYNDAAIELGRELVSRNIDLVYGGGSIGLMGLISHAVYEGGR 181
Query: 71 HVIGVIPKTLMPRELTGETVGEVKAVADMHQRKAEMAKHSDAFIALPGGYGTLEELLEVI 130
HV GVIPKTLMPRELTGETVGEVKAVA+MHQRKAEMAK SDAFIALPGGYGTLEELLEVI
Sbjct: 182 HVTGVIPKTLMPRELTGETVGEVKAVANMHQRKAEMAKRSDAFIALPGGYGTLEELLEVI 361
Query: 131 TWAQLGIHDKPVGLVNVDGYFNSLLSFIDKAVEEGFISPNARHIIVSAPTAKELVKKLEE 190
TWAQLGIHDKPVGL+NVDGY+N+ LSFIDKAVEEGFISP ARHIIVSAPT KELVK++EE
Sbjct: 362 TWAQLGIHDKPVGLLNVDGYYNTFLSFIDKAVEEGFISPTARHIIVSAPTPKELVKEMEE 541
Query: 191 YVPCHEGVASKLSWQMEQ 208
Y P HE V SKLSW+ EQ
Sbjct: 542 YFPQHERVVSKLSWESEQ 595
>TC218844 weakly similar to GB|AAS56573.1|45270384|AY558247 YJL055W
{Saccharomyces cerevisiae;} , partial (37%)
Length = 1027
Score = 329 bits (843), Expect = 7e-91
Identities = 160/210 (76%), Positives = 187/210 (88%)
Frame = +3
Query: 11 SKFKRICVFCGSSPGKKTTYQDAAMNLGNELVSRNIDLVYGGGSIGLMGLVSQAVHDGGR 70
S+F+RICV+CGSSPGK +YQ AA+ LG +LV RNIDLVYGGGSIGLMGL+SQ V+DGGR
Sbjct: 39 SRFRRICVYCGSSPGKNPSYQLAAIQLGKQLVERNIDLVYGGGSIGLMGLISQVVYDGGR 218
Query: 71 HVIGVIPKTLMPRELTGETVGEVKAVADMHQRKAEMAKHSDAFIALPGGYGTLEELLEVI 130
HV+GVIPKTL +E+TGE+VGEV+AV+ MHQRKAEMA+ +DAFIALPGGYGTLEELLE+I
Sbjct: 219 HVLGVIPKTLNAKEITGESVGEVRAVSGMHQRKAEMARQADAFIALPGGYGTLEELLEII 398
Query: 131 TWAQLGIHDKPVGLVNVDGYFNSLLSFIDKAVEEGFISPNARHIIVSAPTAKELVKKLEE 190
TWAQLGIHDKPVGL+NVDGY+NSLL+F+DKAV+EGF++P ARHIIVSA TA+EL+ KLEE
Sbjct: 399 TWAQLGIHDKPVGLLNVDGYYNSLLAFMDKAVDEGFVTPAARHIIVSAHTAQELMCKLEE 578
Query: 191 YVPCHEGVASKLSWQMEQQLAYPQDYDISR 220
YVP H GVA KLSW+MEQQL DISR
Sbjct: 579 YVPEHCGVAPKLSWEMEQQLVNTAKSDISR 668
>BE210785
Length = 385
Score = 229 bits (583), Expect = 1e-60
Identities = 115/116 (99%), Positives = 115/116 (99%)
Frame = +3
Query: 42 VSRNIDLVYGGGSIGLMGLVSQAVHDGGRHVIGVIPKTLMPRELTGETVGEVKAVADMHQ 101
VSRNIDLVYGGGSIGLMGLVSQAVHDGGRHVIGVIPKTLMPRELTGETVGEVKAVADMHQ
Sbjct: 36 VSRNIDLVYGGGSIGLMGLVSQAVHDGGRHVIGVIPKTLMPRELTGETVGEVKAVADMHQ 215
Query: 102 RKAEMAKHSDAFIALPGGYGTLEELLEVITWAQLGIHDKPVGLVNVDGYFNSLLSF 157
RKAEMAKHSDAFIALPGGYGTLEELLEVIT AQLGIHDKPVGLVNVDGYFNSLLSF
Sbjct: 216 RKAEMAKHSDAFIALPGGYGTLEELLEVITXAQLGIHDKPVGLVNVDGYFNSLLSF 383
>BI425016 similar to GP|20197602|gb expressed protein {Arabidopsis thaliana},
partial (60%)
Length = 420
Score = 202 bits (513), Expect = 1e-52
Identities = 96/138 (69%), Positives = 116/138 (83%)
Frame = +2
Query: 76 IPKTLMPRELTGETVGEVKAVADMHQRKAEMAKHSDAFIALPGGYGTLEELLEVITWAQL 135
IPK L+P E++GET GEVK VA+MH+RK+ MAKH+DAFIALPGGYGT+EELLEVI W+QL
Sbjct: 2 IPKALLPLEISGETFGEVKTVANMHERKSVMAKHADAFIALPGGYGTMEELLEVIAWSQL 181
Query: 136 GIHDKPVGLVNVDGYFNSLLSFIDKAVEEGFISPNARHIIVSAPTAKELVKKLEEYVPCH 195
GIHDKPVGL+NVDGYF+SLLS DK VEEGFI +ARHI+V A TA+EL+K++EEYVP H
Sbjct: 182 GIHDKPVGLLNVDGYFHSLLSLFDKGVEEGFIDNSARHIVVIADTAEELIKRMEEYVPNH 361
Query: 196 EGVASKLSWQMEQQLAYP 213
VA++ SW +Q L P
Sbjct: 362 HKVATRQSWARDQLLFEP 415
>BU550433 similar to GP|28372900|gb At5g06300 {Arabidopsis thaliana}, partial
(74%)
Length = 644
Score = 202 bits (513), Expect = 1e-52
Identities = 96/133 (72%), Positives = 116/133 (87%)
Frame = -3
Query: 82 PRELTGETVGEVKAVADMHQRKAEMAKHSDAFIALPGGYGTLEELLEVITWAQLGIHDKP 141
PRE+TG+ +GEV+AV+DMHQRKAEMA+ +DAFIALPGGYGTLEELLE+ITWAQLGIH KP
Sbjct: 642 PREITGDPIGEVRAVSDMHQRKAEMARQADAFIALPGGYGTLEELLEIITWAQLGIHSKP 463
Query: 142 VGLVNVDGYFNSLLSFIDKAVEEGFISPNARHIIVSAPTAKELVKKLEEYVPCHEGVASK 201
VGL+NV+G++NSLLSFIDKAV+EGFISP AR IIVSAPTAK+LV++LEE+VP + V SK
Sbjct: 462 VGLLNVEGFYNSLLSFIDKAVDEGFISPKARRIIVSAPTAKDLVRELEEHVPERDEVVSK 283
Query: 202 LSWQMEQQLAYPQ 214
L W+ P+
Sbjct: 282 LVWEDRLNYVVPE 244
>TC219672 similar to UP|Q9ASW6 (Q9ASW6) T3B23.2/T3B23.2, partial (58%)
Length = 621
Score = 200 bits (509), Expect = 4e-52
Identities = 99/125 (79%), Positives = 113/125 (90%)
Frame = +1
Query: 86 TGETVGEVKAVADMHQRKAEMAKHSDAFIALPGGYGTLEELLEVITWAQLGIHDKPVGLV 145
T E+VGEV+AV+ MHQRKAEMA+ +DAFIALPGGYGTLEELLEVITWAQLGIHDKPVGL+
Sbjct: 4 TIESVGEVRAVSGMHQRKAEMARQADAFIALPGGYGTLEELLEVITWAQLGIHDKPVGLL 183
Query: 146 NVDGYFNSLLSFIDKAVEEGFISPNARHIIVSAPTAKELVKKLEEYVPCHEGVASKLSWQ 205
NVDGY+NSLLSF+D AV+EGFI+P ARHIIVSA TA++L+ KLEEYVP H GVA K SW+
Sbjct: 184 NVDGYYNSLLSFMDNAVDEGFITPAARHIIVSAQTAQDLMCKLEEYVPKHCGVAPKQSWE 363
Query: 206 MEQQL 210
M QQL
Sbjct: 364 MNQQL 378
>TC211902 similar to UP|Q84MC2 (Q84MC2) At5g11950, partial (51%)
Length = 478
Score = 168 bits (425), Expect = 2e-42
Identities = 76/109 (69%), Positives = 94/109 (85%)
Frame = +1
Query: 11 SKFKRICVFCGSSPGKKTTYQDAAMNLGNELVSRNIDLVYGGGSIGLMGLVSQAVHDGGR 70
SKFK +CVFCGS+ G + + DAA+ LGNELV RNIDLVYGGGS+GLMGL+SQ V+DGG
Sbjct: 151 SKFKTVCVFCGSNSGNRQVFSDAAIQLGNELVKRNIDLVYGGGSVGLMGLISQRVYDGGC 330
Query: 71 HVIGVIPKTLMPRELTGETVGEVKAVADMHQRKAEMAKHSDAFIALPGG 119
HV+G+IPK LMP E++GETVGEV+ V+DMH+RKA MA+ +DAF+ALPGG
Sbjct: 331 HVLGIIPKALMPLEISGETVGEVRIVSDMHERKAAMAQEADAFVALPGG 477
>BE474062 similar to GP|9758410|dbj lysine decarboxylase-like protein
{Arabidopsis thaliana}, partial (46%)
Length = 409
Score = 94.0 bits (232), Expect(2) = 3e-35
Identities = 44/61 (72%), Positives = 53/61 (86%)
Frame = +2
Query: 53 GSIGLMGLVSQAVHDGGRHVIGVIPKTLMPRELTGETVGEVKAVADMHQRKAEMAKHSDA 112
G + LMGL+SQ V DGGRHV+GVIP TLMP E+TGE+VGEV+AV+ MHQRKAEMA+ +DA
Sbjct: 227 GVLXLMGLISQVVFDGGRHVLGVIPTTLMPIEITGESVGEVRAVSGMHQRKAEMAREADA 406
Query: 113 F 113
F
Sbjct: 407 F 409
Score = 71.6 bits (174), Expect(2) = 3e-35
Identities = 33/52 (63%), Positives = 41/52 (78%)
Frame = +1
Query: 8 IKLSKFKRICVFCGSSPGKKTTYQDAAMNLGNELVSRNIDLVYGGGSIGLMG 59
+K S+F+RICVFC +S GK +YQ AA+ L +LV RNIDLVYGGGSIG+ G
Sbjct: 91 MKSSRFRRICVFCDTSHGKNPSYQHAAIQLAKQLVERNIDLVYGGGSIGVDG 246
>CD393704
Length = 421
Score = 139 bits (351), Expect = 8e-34
Identities = 67/70 (95%), Positives = 70/70 (99%)
Frame = -1
Query: 151 FNSLLSFIDKAVEEGFISPNARHIIVSAPTAKELVKKLEEYVPCHEGVASKLSWQMEQQL 210
FNSLLSFIDKAVEEGFISPNARHIIVSAPTAKELVKKLE+YVPCHEGVASKLSWQ+EQQL
Sbjct: 412 FNSLLSFIDKAVEEGFISPNARHIIVSAPTAKELVKKLEDYVPCHEGVASKLSWQIEQQL 233
Query: 211 AYPQDYDISR 220
AYPQDYD+SR
Sbjct: 232 AYPQDYDMSR 203
>TC232484 similar to UP|Q9FNH8 (Q9FNH8) Lysine decarboxylase-like protein,
partial (38%)
Length = 450
Score = 138 bits (347), Expect = 2e-33
Identities = 64/88 (72%), Positives = 77/88 (86%)
Frame = +2
Query: 11 SKFKRICVFCGSSPGKKTTYQDAAMNLGNELVSRNIDLVYGGGSIGLMGLVSQAVHDGGR 70
S+F+RICVFCG+SPGK +YQ AA+ L +LV RNIDLVYGGGSIGLMGL+SQ V DGGR
Sbjct: 185 SRFRRICVFCGTSPGKNPSYQLAAIQLAKQLVERNIDLVYGGGSIGLMGLISQVVFDGGR 364
Query: 71 HVIGVIPKTLMPRELTGETVGEVKAVAD 98
HV+GVIP TLMPRE+TGE+VGEV++V +
Sbjct: 365 HVLGVIPTTLMPREITGESVGEVESVGE 448
>TC209497 similar to UP|Q9ASW6 (Q9ASW6) T3B23.2/T3B23.2, partial (41%)
Length = 464
Score = 136 bits (342), Expect = 9e-33
Identities = 68/88 (77%), Positives = 77/88 (87%)
Frame = -1
Query: 133 AQLGIHDKPVGLVNVDGYFNSLLSFIDKAVEEGFISPNARHIIVSAPTAKELVKKLEEYV 192
AQLGIHDKPVGL+NVDGY+NSLLSFIDKAVEEGFISP ARHIIVSAP+ KELVK++EEY
Sbjct: 464 AQLGIHDKPVGLLNVDGYYNSLLSFIDKAVEEGFISPKARHIIVSAPSTKELVKEMEEYF 285
Query: 193 PCHEGVASKLSWQMEQQLAYPQDYDISR 220
P HE VASKLSW+ E Q+ Y + D+SR
Sbjct: 284 PQHERVASKLSWETE-QIDYSSNCDMSR 204
>BQ453944 similar to GP|20804658|dbj lysine decarboxylase-like protein {Oryza
sativa (japonica cultivar-group)}, partial (39%)
Length = 368
Score = 103 bits (257), Expect(2) = 4e-29
Identities = 47/60 (78%), Positives = 54/60 (89%)
Frame = +3
Query: 108 KHSDAFIALPGGYGTLEELLEVITWAQLGIHDKPVGLVNVDGYFNSLLSFIDKAVEEGFI 167
+ +DAFIALPGGYGT+EELLE+ITWAQLGIH KPVGL+NVDGY+NSLL+ D VEEGFI
Sbjct: 3 QEADAFIALPGGYGTMEELLEMITWAQLGIHKKPVGLLNVDGYYNSLLALFDNGVEEGFI 182
Score = 41.6 bits (96), Expect(2) = 4e-29
Identities = 19/36 (52%), Positives = 26/36 (71%)
Frame = +1
Query: 173 HIIVSAPTAKELVKKLEEYVPCHEGVASKLSWQMEQ 208
+I+V+A +AKEL+ K+E+Y P HE VA SWQ Q
Sbjct: 199 NILVAASSAKELMMKMEQYSPSHEHVAPHDSWQTRQ 306
>BM270992 similar to GP|9758410|dbj| lysine decarboxylase-like protein
{Arabidopsis thaliana}, partial (43%)
Length = 467
Score = 99.4 bits (246), Expect(2) = 7e-29
Identities = 46/64 (71%), Positives = 55/64 (85%)
Frame = +1
Query: 11 SKFKRICVFCGSSPGKKTTYQDAAMNLGNELVSRNIDLVYGGGSIGLMGLVSQAVHDGGR 70
S+F+RICV+CGSSPGK +YQ AA+ LG +LV IDLVYGGGSIGLMGL+SQ V+DGGR
Sbjct: 118 SRFRRICVYCGSSPGKNPSYQLAAIQLGKQLVV*YIDLVYGGGSIGLMGLISQVVYDGGR 297
Query: 71 HVIG 74
HV+G
Sbjct: 298 HVLG 309
Score = 45.1 bits (105), Expect(2) = 7e-29
Identities = 20/29 (68%), Positives = 26/29 (88%)
Frame = +3
Query: 75 VIPKTLMPRELTGETVGEVKAVADMHQRK 103
VIP+TL RE+TGE+VGE++AV+ MHQRK
Sbjct: 381 VIPETLNAREITGESVGELRAVSGMHQRK 467
>AW706971 similar to GP|13605607|gb| T3B23.2/T3B23.2 {Arabidopsis thaliana},
partial (31%)
Length = 422
Score = 110 bits (276), Expect = 4e-25
Identities = 53/67 (79%), Positives = 59/67 (87%)
Frame = +3
Query: 142 VGLVNVDGYFNSLLSFIDKAVEEGFISPNARHIIVSAPTAKELVKKLEEYVPCHEGVASK 201
VGL+NVDGY+N+ LSFIDKAVEEGFISP ARHIIVSAPT KELVK++EEY P HE V SK
Sbjct: 87 VGLLNVDGYYNTFLSFIDKAVEEGFISPTARHIIVSAPTPKELVKEMEEYFPQHERVVSK 266
Query: 202 LSWQMEQ 208
LSW+ EQ
Sbjct: 267 LSWESEQ 287
>AI973691 similar to GP|28372900|gb| At5g06300 {Arabidopsis thaliana},
partial (40%)
Length = 199
Score = 109 bits (272), Expect = 1e-24
Identities = 53/66 (80%), Positives = 56/66 (84%)
Frame = +2
Query: 111 DAFIALPGGYGTLEELLEVITWAQLGIHDKPVGLVNVDGYFNSLLSFIDKAVEEGFISPN 170
DAFI LPGGYGTLEELLEVITWAQLGIH KP GL+N DG +NSLL IDKAV+EG ISP
Sbjct: 2 DAFIVLPGGYGTLEELLEVITWAQLGIHSKPAGLLNADGCYNSLLPLIDKAVDEGSISPT 181
Query: 171 ARHIIV 176
AR IIV
Sbjct: 182 ARRIIV 199
>BU544251
Length = 423
Score = 94.0 bits (232), Expect = 5e-20
Identities = 47/55 (85%), Positives = 50/55 (90%), Gaps = 1/55 (1%)
Frame = -1
Query: 142 VGLVNVDGYFNSLLSFIDKAVEEGFISPNARHIIVSAPTAKELVKKLEE-YVPCH 195
VGLVNVDGYFNSLLSFIDKAVEEGFISPNARHIIVS PT+KELVKK E ++ CH
Sbjct: 420 VGLVNVDGYFNSLLSFIDKAVEEGFISPNARHIIVSXPTSKELVKKXEVIFIGCH 256
Score = 62.0 bits (149), Expect = 2e-10
Identities = 25/32 (78%), Positives = 30/32 (93%)
Frame = -3
Query: 189 EEYVPCHEGVASKLSWQMEQQLAYPQDYDISR 220
++YVPCHE VASKLSWQ+EQQL YP++YDISR
Sbjct: 130 QDYVPCHESVASKLSWQIEQQLTYPEEYDISR 35
>CD407938 homologue to PIR|S04125|S041 chlorophyll a/b-binding protein type
III precursor - tomato, partial (18%)
Length = 689
Score = 74.3 bits (181), Expect = 4e-14
Identities = 32/36 (88%), Positives = 36/36 (99%)
Frame = -3
Query: 185 VKKLEEYVPCHEGVASKLSWQMEQQLAYPQDYDISR 220
+KKLE+YVPCHEGVASKLSWQ+EQQLAYPQDYD+SR
Sbjct: 345 LKKLEDYVPCHEGVASKLSWQIEQQLAYPQDYDMSR 238
>CD413732 homologue to PIR|E96640|E966 PSI type III chlorophyll a/b-binding
protein [imported] - Arabidopsis thaliana, partial (7%)
Length = 605
Score = 74.3 bits (181), Expect = 4e-14
Identities = 32/36 (88%), Positives = 36/36 (99%)
Frame = -3
Query: 185 VKKLEEYVPCHEGVASKLSWQMEQQLAYPQDYDISR 220
+KKLE+YVPCHEGVASKLSWQ+EQQLAYPQDYD+SR
Sbjct: 354 LKKLEDYVPCHEGVASKLSWQIEQQLAYPQDYDMSR 247
>BM887010
Length = 421
Score = 68.9 bits (167), Expect = 2e-12
Identities = 33/37 (89%), Positives = 35/37 (94%)
Frame = +2
Query: 84 ELTGETVGEVKAVADMHQRKAEMAKHSDAFIALPGGY 120
+LTGETVGEVKAVA+MHQRKAEMAK SDAFIALPG Y
Sbjct: 50 QLTGETVGEVKAVANMHQRKAEMAKRSDAFIALPGSY 160
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,781,319
Number of Sequences: 63676
Number of extensions: 101204
Number of successful extensions: 568
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 568
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 568
length of query: 220
length of database: 12,639,632
effective HSP length: 93
effective length of query: 127
effective length of database: 6,717,764
effective search space: 853156028
effective search space used: 853156028
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC148995.1


