

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148994.4 - phase: 0
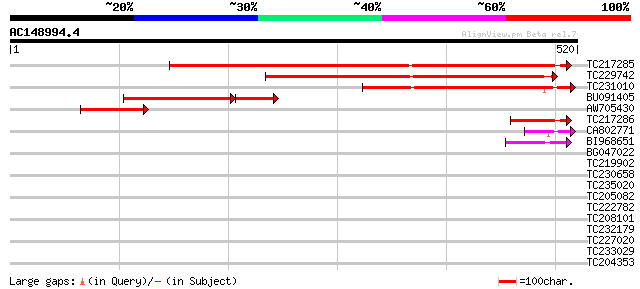
(520 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC217285 similar to UP|Q8H9B4 (Q8H9B4) UDP-glucose:sterol 3-O-gl... 582 e-166
TC229742 weakly similar to UP|Q8H9B4 (Q8H9B4) UDP-glucose:sterol... 340 7e-94
TC231010 similar to UP|Q8H9B4 (Q8H9B4) UDP-glucose:sterol 3-O-gl... 323 1e-88
BU091405 151 2e-46
AW705430 homologue to GP|24459977|dbj UDP-glucose:sterol 3-O-glu... 108 8e-24
TC217286 weakly similar to UP|Q8H9B4 (Q8H9B4) UDP-glucose:sterol... 65 1e-10
CA802771 57 2e-08
BI968651 45 6e-05
BG047022 40 0.003
TC219902 similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransferase H... 33 0.40
TC230658 similar to UP|Q9FT43 (Q9FT43) Sterol glucosyltransferas... 32 0.52
TC235020 31 1.2
TC205082 weakly similar to UP|Q8GT67 (Q8GT67) Xyloglucan-specifi... 30 2.6
TC222782 30 2.6
TC208101 weakly similar to UP|Q9ZWS2 (Q9ZWS2) Flavonoid 3-O-gala... 30 3.4
TC232179 weakly similar to UP|Q6K5X7 (Q6K5X7) Transport protein-... 29 4.4
TC227020 similar to UP|Q8L6S5 (Q8L6S5) IAA16 protein, partial (63%) 29 5.8
TC233029 homologue to GB|AAN46837.1|24111405|BT001072 At5g62000/... 28 7.6
TC204353 homologue to UP|Q8S3F8 (Q8S3F8) S-adenosylmethionine de... 28 7.6
>TC217285 similar to UP|Q8H9B4 (Q8H9B4) UDP-glucose:sterol
3-O-glucosyltransferase, partial (59%)
Length = 1302
Score = 582 bits (1500), Expect = e-166
Identities = 272/369 (73%), Positives = 315/369 (84%)
Frame = +2
Query: 147 QRSQIRAIIHSLLPACNSRYPESNEPFKADAIIANPPAYGHTHVAEYLNVPLHIFFTMPW 206
QR+Q++ II+SLLPAC +S PFKADAIIANPPAYGHTHVAE L +P+HIFFTMPW
Sbjct: 2 QRNQMKEIINSLLPACKEPDIDSGVPFKADAIIANPPAYGHTHVAEALKIPIHIFFTMPW 181
Query: 207 TPTSDFPHPLSRVRQPIGYRLSYQIVDALIWLGIRDLINEFRKKKLKLRAVTYLRGSYTF 266
TPT++FPHPLSRV+Q GYRLSYQIVD+LIWLGIRD+IN+ RKKKLKLR VTYL GS
Sbjct: 182 TPTTEFPHPLSRVKQQAGYRLSYQIVDSLIWLGIRDMINDLRKKKLKLRPVTYLSGSQGS 361
Query: 267 PPDMPYGYIWSPHLVPKPKDWGPNIDIVGFCFLDLASNYEPPKSLVDWLEEGENPIYVGF 326
D+P+ YIWSPHLVPKPKDWGP ID+VGFCFLDLA NYEPP+SLV WLEEG+ PIY+GF
Sbjct: 362 ETDVPHAYIWSPHLVPKPKDWGPKIDVVGFCFLDLALNYEPPESLVKWLEEGDKPIYIGF 541
Query: 327 GSLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGLGNLAELNTSKSVYLLDNCPHDWLFP 386
GSLP+QEP+KMT+IIV ALE TGQRGIINKGWGGLGNLAE S+YLLDNCPHDWLF
Sbjct: 542 GSLPVQEPKKMTQIIVDALEITGQRGIINKGWGGLGNLAE--PKDSIYLLDNCPHDWLFL 715
Query: 387 RCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPFWGERVHARGVGPAPIRVEEFTLERL 446
RC AVVHHGGAGTTAAGL+A CPTT+VPFFGDQPFWGERVHARGVGP PI V+EF+L +L
Sbjct: 716 RCKAVVHHGGAGTTAAGLKAACPTTIVPFFGDQPFWGERVHARGVGPPPIPVDEFSLPKL 895
Query: 447 VDAIRFMLNPEVKKRAVELANAMKNEDGVAGAVNAFYKHYPREKPDTEAEPRPVPSVHKH 506
VDAI+ ML+P+VK+RA+ELA AM+NEDGV GAV AF+K P++K +++A+P+P
Sbjct: 896 VDAIKLMLDPKVKERAIELAKAMENEDGVTGAVKAFFKQLPQKKSESDADPQPT----GF 1063
Query: 507 LSIRGCFGC 515
S+R CFGC
Sbjct: 1064FSVRRCFGC 1090
>TC229742 weakly similar to UP|Q8H9B4 (Q8H9B4) UDP-glucose:sterol
3-O-glucosyltransferase, partial (34%)
Length = 1133
Score = 340 bits (873), Expect = 7e-94
Identities = 150/268 (55%), Positives = 206/268 (75%)
Frame = +2
Query: 235 LIWLGIRDLINEFRKKKLKLRAVTYLRGSYTFPPDMPYGYIWSPHLVPKPKDWGPNIDIV 294
LIW GIR +IN+FRK+KLKL + Y +P GY+WSPH+VPKP DWGP +D+V
Sbjct: 14 LIWWGIRGIINDFRKRKLKLAPIAYFSMYSGSISHLPTGYMWSPHVVPKPSDWGPLVDVV 193
Query: 295 GFCFLDLASNYEPPKSLVDWLEEGENPIYVGFGSLPLQEPEKMTRIIVQALEQTGQRGII 354
G+CFL+L S Y+P + V W+++G P+Y GFGS+PL +P++ T +IV+AL+ TGQRGII
Sbjct: 194 GYCFLNLGSKYQPQEDFVRWIQKGPKPLYFGFGSMPLDDPKRTTDVIVEALKDTGQRGII 373
Query: 355 NKGWGGLGNLAELNTSKSVYLLDNCPHDWLFPRCAAVVHHGGAGTTAAGLRAECPTTVVP 414
++GWG LGNLAE+ +V++L+ CPHDWLFP+C+A+VHHGGAGTTA GL+A CPTT+VP
Sbjct: 374 DRGWGNLGNLAEV--PDNVFVLEECPHDWLFPQCSALVHHGGAGTTATGLKAGCPTTIVP 547
Query: 415 FFGDQPFWGERVHARGVGPAPIRVEEFTLERLVDAIRFMLNPEVKKRAVELANAMKNEDG 474
FFGDQ FWG+R++ +G+GPAPI + + +LE L ++I+FML PEVK RA+E+A ++NEDG
Sbjct: 548 FFGDQFFWGDRINQKGLGPAPIPISQLSLENLSNSIKFMLQPEVKSRAMEVAKLIENEDG 727
Query: 475 VAGAVNAFYKHYPREKPDTEAEPRPVPS 502
V AV++F++H P E P P+PS
Sbjct: 728 VTAAVDSFHRHLPPEL------PLPIPS 793
>TC231010 similar to UP|Q8H9B4 (Q8H9B4) UDP-glucose:sterol
3-O-glucosyltransferase, partial (28%)
Length = 893
Score = 323 bits (828), Expect = 1e-88
Identities = 158/198 (79%), Positives = 174/198 (87%), Gaps = 2/198 (1%)
Frame = +1
Query: 324 VGFGSLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGLGNLAELNTSKSVYLLDNCPHDW 383
VGFGSLPLQ+PEK+T+II+QALE+TGQRGIINKGWGGLG+LAE N KSVYLLDNCPHDW
Sbjct: 1 VGFGSLPLQQPEKITQIIIQALEETGQRGIINKGWGGLGSLAEQN--KSVYLLDNCPHDW 174
Query: 384 LFPRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPFWGERVHARGVGPAPIRVEEFTL 443
LFPRC AVVHHGGAGTTAAGLRAECPTT+VPFFGDQPFWG+RV ARGVGPAPI V+EF+
Sbjct: 175 LFPRCTAVVHHGGAGTTAAGLRAECPTTIVPFFGDQPFWGDRVRARGVGPAPIPVDEFSF 354
Query: 444 ERLVDAIRFMLNPEVKKRAVELANAMKNEDGVAGAVNAFYKHYPRE--KPDTEAEPRPVP 501
+RLVDAI ML PEVKKRAVELANAMKNE+GV GAV AFYKHYP E + + AEP+
Sbjct: 355 DRLVDAIHLMLKPEVKKRAVELANAMKNENGVLGAVKAFYKHYPAEFKRLASTAEPK--- 525
Query: 502 SVHKHLSIRGCFGCYHSS 519
SVHK+ SIRGCFGC SS
Sbjct: 526 SVHKYFSIRGCFGCSKSS 579
>BU091405
Length = 431
Score = 151 bits (381), Expect(2) = 2e-46
Identities = 71/103 (68%), Positives = 83/103 (79%)
Frame = +1
Query: 105 FEDFVLSAGLEFYPLGGDPKVLAEYMVKNKGFLPSGPSEIHLQRSQIRAIIHSLLPACNS 164
F+ FV SAG+ FYPLGGDP+ LAEYM +NKG +PSGP+EI +QR Q++AII SLLPAC S
Sbjct: 1 FDTFVKSAGVNFYPLGGDPRALAEYMARNKGIIPSGPTEISIQRKQLKAIIDSLLPACIS 180
Query: 165 RYPESNEPFKADAIIANPPAYGHTHVAEYLNVPLHIFFTMPWT 207
E+ PF+A AII+NP A GHTHVAE L VPLHIFFTMPWT
Sbjct: 181 PDLETGVPFRAQAIISNPTACGHTHVAEALGVPLHIFFTMPWT 309
Score = 53.5 bits (127), Expect(2) = 2e-46
Identities = 25/39 (64%), Positives = 29/39 (74%)
Frame = +2
Query: 208 PTSDFPHPLSRVRQPIGYRLSYQIVDALIWLGIRDLINE 246
PT +F HPL+RV Q GY LSY IVD LIW GIR +IN+
Sbjct: 314 PTYEFSHPLARVPQSAGYWLSYIIVDLLIWWGIRGIIND 430
>AW705430 homologue to GP|24459977|dbj UDP-glucose:sterol
3-O-glucosyltransferase {Panax ginseng}, partial (10%)
Length = 433
Score = 108 bits (269), Expect = 8e-24
Identities = 51/62 (82%), Positives = 56/62 (90%)
Frame = +2
Query: 66 PQQIAMLIVGTRGDVQPFVAIGKRLQADGHRVRLATHKNFEDFVLSAGLEFYPLGGDPKV 125
P I MLIVGTRGDVQPF+AIGKR+Q GHRVRLATH NF++FVL+AGLEFYPLGGDPKV
Sbjct: 2 PLNIVMLIVGTRGDVQPFIAIGKRMQDYGHRVRLATHSNFKEFVLTAGLEFYPLGGDPKV 181
Query: 126 LA 127
LA
Sbjct: 182 LA 187
>TC217286 weakly similar to UP|Q8H9B4 (Q8H9B4) UDP-glucose:sterol
3-O-glucosyltransferase, partial (7%)
Length = 436
Score = 64.7 bits (156), Expect = 1e-10
Identities = 29/56 (51%), Positives = 40/56 (70%)
Frame = +2
Query: 460 KRAVELANAMKNEDGVAGAVNAFYKHYPREKPDTEAEPRPVPSVHKHLSIRGCFGC 515
+RA+ELA AM+NEDGV GAV AF+K P++KP+ +A+P+P S+ CFGC
Sbjct: 2 ERAIELAKAMENEDGVTGAVKAFFKQLPQKKPEPDADPQPT----SFFSVGRCFGC 157
>CA802771
Length = 456
Score = 57.4 bits (137), Expect = 2e-08
Identities = 31/57 (54%), Positives = 34/57 (59%), Gaps = 10/57 (17%)
Frame = +1
Query: 473 DGVAGAVNAFYKHYPREKPD----------TEAEPRPVPSVHKHLSIRGCFGCYHSS 519
DGV G V AFYKHYP EK + AEP+P VHK+ SIRGCFGC SS
Sbjct: 1 DGVLGLVKAFYKHYPPEKSKFDDAKSKQLASTAEPKP---VHKYFSIRGCFGCSKSS 162
>BI968651
Length = 349
Score = 45.4 bits (106), Expect = 6e-05
Identities = 25/61 (40%), Positives = 36/61 (58%)
Frame = -1
Query: 455 NPEVKKRAVELANAMKNEDGVAGAVNAFYKHYPREKPDTEAEPRPVPSVHKHLSIRGCFG 514
+P+VK RA E+A + NED V+G V AF + P++KP EP ++ LS+ FG
Sbjct: 349 DPKVK*RASEVAKSXXNEDXVSGPVKAFGQQLPQKKP----EPDTNAQTYRSLSVGRGFG 182
Query: 515 C 515
C
Sbjct: 181 C 179
>BG047022
Length = 405
Score = 40.0 bits (92), Expect = 0.003
Identities = 14/20 (70%), Positives = 18/20 (90%)
Frame = +1
Query: 408 CPTTVVPFFGDQPFWGERVH 427
CPTT+VPFFGDQ FW +R++
Sbjct: 334 CPTTIVPFFGDQFFWRDRIY 393
>TC219902 similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransferase HRA25,
partial (52%)
Length = 967
Score = 32.7 bits (73), Expect = 0.40
Identities = 47/193 (24%), Positives = 75/193 (38%), Gaps = 20/193 (10%)
Frame = +1
Query: 305 YEPPKSLVDWLEEGENP--IYVGFGSLPLQEPEKMTRI----------IVQALEQTGQRG 352
+E S + WL++ + +YV FGS + + + + + Q +R
Sbjct: 163 WEEDLSCMSWLDQQPHGSVLYVAFGSFTHFDQNQFNELALGIDLTNRPFLWVVRQDNKRV 342
Query: 353 IINKGWGGLGNLAELNTSKSVYLLDNCPHDWLFPRCAAVVHHGGAGTTAAGLRAECPTTV 412
N+ G G + + V P A V H G +T G+ P
Sbjct: 343 YPNEFLGCKGKIVSWAPQQKVLS---------HPAIACFVTHCGWNSTIEGVSNGLPLLC 495
Query: 413 VPFFGDQ----PFWGERVHARGVGPAPIR---VEEFTLERLVDAIRFMLNPE-VKKRAVE 464
P+FGDQ + + + G+G + V LER VD I LN E +K R++E
Sbjct: 496 WPYFGDQICNKTYICDELKV-GLGFDSDKNGLVSRMELERKVDQI---LNDENIKSRSLE 663
Query: 465 LANAMKNEDGVAG 477
L + + N AG
Sbjct: 664 LKDKVMNNIAKAG 702
>TC230658 similar to UP|Q9FT43 (Q9FT43) Sterol glucosyltransferase-like
protein, partial (6%)
Length = 883
Score = 32.3 bits (72), Expect = 0.52
Identities = 13/25 (52%), Positives = 16/25 (64%)
Frame = +3
Query: 412 VVPFFGDQPFWGERVHARGVGPAPI 436
V PF DQ +W ER+H GV P P+
Sbjct: 732 VCPFILDQFYWAERMHWLGVSPEPL 806
>TC235020
Length = 447
Score = 31.2 bits (69), Expect = 1.2
Identities = 20/58 (34%), Positives = 30/58 (51%), Gaps = 2/58 (3%)
Frame = -2
Query: 219 VRQPIGYRLSYQIVDALIWLGIRDLINEFRKKKLKLRAVTYLRGSYTFPP--DMPYGY 274
+++ +G +Y I L W GI + + RKKKLKL V + TFP +P+ Y
Sbjct: 350 IKEKLGSNNNYYIFVKLFWYGIPLIKQKKRKKKLKLEEVETDL*T*TFPKGLSIPWRY 177
>TC205082 weakly similar to UP|Q8GT67 (Q8GT67) Xyloglucan-specific fungal
endoglucanase inhibitor protein, partial (25%)
Length = 1773
Score = 30.0 bits (66), Expect = 2.6
Identities = 21/69 (30%), Positives = 29/69 (41%), Gaps = 2/69 (2%)
Frame = +2
Query: 151 IRAIIHSLLPACNSRYPESNEPFKADAIIANPPAYG--HTHVAEYLNVPLHIFFTMPWTP 208
I +++S LP C R P +P I P A H + +P + T+PW P
Sbjct: 116 ILPLLYS*LP-CLIR-PSHTKPNPTQPTIFQPAAATKQRPHPQNHFQLPTTLSITLPWLP 289
Query: 209 TSDFPHPLS 217
F H LS
Sbjct: 290 LPSFSHSLS 316
>TC222782
Length = 417
Score = 30.0 bits (66), Expect = 2.6
Identities = 11/24 (45%), Positives = 16/24 (65%)
Frame = -3
Query: 496 EPRPVPSVHKHLSIRGCFGCYHSS 519
+P VPS +H R CFGC++S+
Sbjct: 106 DPSEVPSASQHGEFRYCFGCWNSA 35
>TC208101 weakly similar to UP|Q9ZWS2 (Q9ZWS2) Flavonoid 3-O-galactosyl
transferase, partial (46%)
Length = 1022
Score = 29.6 bits (65), Expect = 3.4
Identities = 41/177 (23%), Positives = 70/177 (39%), Gaps = 11/177 (6%)
Frame = +3
Query: 312 VDWL--EEGENPIYVGFGSLPLQEPEKMTRIIVQALEQTG-------QRGIINKGWGGLG 362
+ WL + ++ YV FG++ P ++ + +ALE++G + G+++ L
Sbjct: 213 LSWLGMKNSKSVAYVCFGTVVAPPPHELVAV-AEALEESGFPFLWSLKEGLMSL----LP 377
Query: 363 NLAELNTSKSVYLLDNCP--HDWLFPRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQP 420
N T K ++ P H V H GA + + + P PFFGDQ
Sbjct: 378 NGFVERTKKRGKIVSWAPQTHVLAHDSVGVFVTHCGANSVIESVSSGVPMICRPFFGDQG 557
Query: 421 FWGERVHARGVGPAPIRVEEFTLERLVDAIRFMLNPEVKKRAVELANAMKNEDGVAG 477
+ I + FT LV ++ +L E K+ + A +K AG
Sbjct: 558 VAARVIEDVWEIGMMIEGKMFTKNGLVKSLNLILVHEEGKKIRDNALRVKKTVEDAG 728
>TC232179 weakly similar to UP|Q6K5X7 (Q6K5X7) Transport protein-like,
partial (4%)
Length = 782
Score = 29.3 bits (64), Expect = 4.4
Identities = 26/106 (24%), Positives = 45/106 (41%), Gaps = 3/106 (2%)
Frame = +1
Query: 399 TTAAGLRAECPTTVVPFFGDQPFWGERVHARGVGPAPIRVEEFTLERLVDAIRFMLNPEV 458
TTA+ AECP T++ R + + + E +++V M NP
Sbjct: 151 TTASSKLAECPETILNL---------RKQLKALASSN---EVAIFDKVVSTTNTMANPTQ 294
Query: 459 KKRAVE---LANAMKNEDGVAGAVNAFYKHYPREKPDTEAEPRPVP 501
KK ++ L N M+ ED G + +K P E+ ++ + + P
Sbjct: 295 KKNLIKRSSLRNQMQAEDEAKGGM---HKSVPTEETKSDKDVQRPP 423
>TC227020 similar to UP|Q8L6S5 (Q8L6S5) IAA16 protein, partial (63%)
Length = 1104
Score = 28.9 bits (63), Expect = 5.8
Identities = 12/26 (46%), Positives = 15/26 (57%)
Frame = -3
Query: 142 SEIHLQRSQIRAIIHSLLPACNSRYP 167
S IH Q+ +HS+LPAC R P
Sbjct: 382 SSIHRHLHQLATALHSILPACPHRRP 305
>TC233029 homologue to GB|AAN46837.1|24111405|BT001072 At5g62000/mtg10_20
{Arabidopsis thaliana;} , partial (23%)
Length = 1004
Score = 28.5 bits (62), Expect = 7.6
Identities = 15/41 (36%), Positives = 23/41 (55%), Gaps = 2/41 (4%)
Frame = -3
Query: 246 EFRKKKLKLRAVT--YLRGSYTFPPDMPYGYIWSPHLVPKP 284
++++KK KL T +LR YTF P + + + PH P P
Sbjct: 903 QWKEKKRKLA*ATKFFLRNHYTFIPLLGSLFTFKPHFEPHP 781
>TC204353 homologue to UP|Q8S3F8 (Q8S3F8) S-adenosylmethionine decarboxylase,
partial (58%)
Length = 1265
Score = 28.5 bits (62), Expect = 7.6
Identities = 13/35 (37%), Positives = 18/35 (51%)
Frame = +3
Query: 238 LGIRDLINEFRKKKLKLRAVTYLRGSYTFPPDMPY 272
L I ++ L +R+V Y RGS+ FP PY
Sbjct: 858 LAIPPILKFAEMLSLNVRSVNYTRGSFIFPGAQPY 962
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.141 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,777,768
Number of Sequences: 63676
Number of extensions: 425440
Number of successful extensions: 1951
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 1904
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1943
length of query: 520
length of database: 12,639,632
effective HSP length: 102
effective length of query: 418
effective length of database: 6,144,680
effective search space: 2568476240
effective search space used: 2568476240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148994.4


