

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148970.6 - phase: 0
(355 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
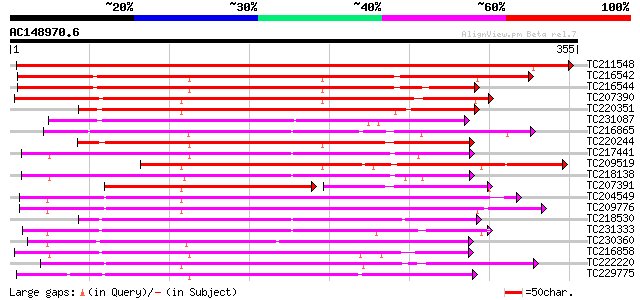
Score E
Sequences producing significant alignments: (bits) Value
TC211548 weakly similar to UP|Q7XIP7 (Q7XIP7) Receptor-like kina... 532 e-151
TC216542 weakly similar to UP|Q9LL55 (Q9LL55) Receptor-like kina... 261 3e-70
TC216544 weakly similar to UP|Q9LL55 (Q9LL55) Receptor-like kina... 241 5e-64
TC207390 weakly similar to UP|Q9ATQ5 (Q9ATQ5) LRK33, partial (34%) 240 6e-64
TC220351 similar to UP|Q961E7 (Q961E7) GH28523p (CG1830-PB) , pa... 217 7e-57
TC231087 similar to UP|Q39203 (Q39203) Receptor-like protein kin... 213 1e-55
TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial... 210 7e-55
TC220244 weakly similar to UP|Q9LL58 (Q9LL58) Receptor-like kina... 204 4e-53
TC217441 homologue to GB|AAK68074.1|14573459|AF384970 somatic em... 200 7e-52
TC209519 similar to UP|Q84K89 (Q84K89) Receptor kinase LRK10 (Re... 195 3e-50
TC218138 homologue to UP|Q8GRK2 (Q8GRK2) Somatic embryogenesis r... 192 3e-49
TC207391 similar to UP|Q9SWU8 (Q9SWU8) Receptor-like kinase (Fra... 149 1e-48
TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-l... 187 5e-48
TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-l... 187 6e-48
TC218530 similar to UP|Q9SGU0 (Q9SGU0) T6H22.8.1 protein, partia... 183 1e-46
TC231333 similar to UP|Q8L741 (Q8L741) AT3g13690/MMM17_12, parti... 179 2e-45
TC230360 similar to GB|AAP68229.1|31711746|BT008790 At5g63940 {A... 175 2e-44
TC216858 homologue to UP|Q76FZ8 (Q76FZ8) Brassinosteroid recepto... 173 1e-43
TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like prot... 172 3e-43
TC229775 weakly similar to UP|KPEL_DROME (Q05652) Probable serin... 171 5e-43
>TC211548 weakly similar to UP|Q7XIP7 (Q7XIP7) Receptor-like kinase
TAK33-like protein, partial (58%)
Length = 1194
Score = 532 bits (1370), Expect = e-151
Identities = 260/353 (73%), Positives = 302/353 (84%), Gaps = 4/353 (1%)
Frame = +2
Query: 5 KPIRFTSQQLRIATDNYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAE 64
KPIRFT QQLRIATDNYS LLGSGGFG VYKG F+NGT+VAVKVLRGSSDK+I+EQFMAE
Sbjct: 2 KPIRFTDQQLRIATDNYSFLLGSGGFGVVYKGSFSNGTIVAVKVLRGSSDKRIDEQFMAE 181
Query: 65 VGTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNALGYEKLHEIAIGT 124
VGTIG++HHFNLV+L+GFCFE++L ALVYEYM NG+L++YL HE L +EKLHEIA+GT
Sbjct: 182 VGTIGKVHHFNLVRLYGFCFERHLRALVYEYMVNGALEKYLFHENTTLSFEKLHEIAVGT 361
Query: 125 ARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTP 184
ARGIAYLHE C+ RI+HYDIKPGNILLD NF PKVADFGLAKLCNR+NTHI+MTGGRGTP
Sbjct: 362 ARGIAYLHEECQQRIIHYDIKPGNILLDRNFCPKVADFGLAKLCNRDNTHISMTGGRGTP 541
Query: 185 GYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVWKKFDAGLL 244
GYAAPELW+PFP+THKCDVYSFGMLLFEI+GRRRN +I ESQ WFP+WVW++FDA +
Sbjct: 542 GYAAPELWLPFPVTHKCDVYSFGMLLFEIIGRRRNHNINLPESQVWFPMWVWERFDAENV 721
Query: 245 EEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLEIPKTFNPFQH 304
E+ + CGIE++NREIAER+VKVAL CVQY+ + RP+MS VVKMLEGS+E+PK NPFQH
Sbjct: 722 EDLISACGIEDQNREIAERIVKVALSCVQYKPEARPIMSVVVKMLEGSVEVPKPLNPFQH 901
Query: 305 LIDETKFTTHSDQESNTYT-TSV---SSVMVSDSNIVCATPIMRKYEIELASS 353
LID T T Q S T T TS+ SS +V+ S V ATP+M KYEIELAS+
Sbjct: 902 LIDWTPPPTDPVQASQTNTDTSICSDSSELVAKSVHVVATPVMTKYEIELASA 1060
>TC216542 weakly similar to UP|Q9LL55 (Q9LL55) Receptor-like kinase, partial
(38%)
Length = 1435
Score = 261 bits (668), Expect = 3e-70
Identities = 146/331 (44%), Positives = 214/331 (64%), Gaps = 8/331 (2%)
Frame = +2
Query: 6 PIRFTSQQLRIATDNYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEV 65
PI ++ ++++ + LG GG+G V+KG ++G VA+K+L + K + F++E+
Sbjct: 368 PIGYSYKEIKKMARGFKEKLGGGGYGIVFKGKLHSGPSVAIKMLHKA--KGNGQDFISEI 541
Query: 66 GTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA--LGYEKLHEIAIG 123
TIGRIHH N+V+L G+C E + ALVYE+M NGSLD+++ + L Y++++ IAIG
Sbjct: 542 ATIGRIHHQNVVQLIGYCVEGSKRALVYEFMPNGSLDKFIFTKDGNIHLTYDEIYNIAIG 721
Query: 124 TARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGT 183
ARGIAYLH CE +I+H+DIKP NILLD F PKV+DFGLAKL +N+ +TMT RGT
Sbjct: 722 VARGIAYLHHGCEMQILHFDIKPHNILLDETFTPKVSDFGLAKLYPIDNSIVTMTAARGT 901
Query: 184 PGYAAPELWMP--FPITHKCDVYSFGMLLFEIVGRRRNLD-IKNTESQEWFPIWVWKKFD 240
GY APEL+ I+HK DVYSFGMLL E+ +R+NL+ + SQ +FP W++ +
Sbjct: 902 IGYMAPELFYKNIGGISHKADVYSFGMLLMEMASKRKNLNPHADHSSQLYFPFWIYNQLG 1081
Query: 241 AGLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEG---SLEIPK 297
E + + G+ E+ +IA++M+ V+LWC+Q + RP M+ VV+MLEG SLEIP
Sbjct: 1082K---ETDIEMEGVTEEENKIAKKMIIVSLWCIQLKPTDRPSMNKVVEMLEGDIESLEIPP 1252
Query: 298 TFNPFQHLIDETKFTTHSDQESNTYTTSVSS 328
+ + H E + +S Q +T S S+
Sbjct: 1253KPSLYPHETMENDQSIYSSQTMSTDFISSSN 1345
>TC216544 weakly similar to UP|Q9LL55 (Q9LL55) Receptor-like kinase, partial
(36%)
Length = 1229
Score = 241 bits (614), Expect = 5e-64
Identities = 131/294 (44%), Positives = 193/294 (65%), Gaps = 5/294 (1%)
Frame = +2
Query: 6 PIRFTSQQLRIATDNYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEV 65
PIR++ ++++ + + LG GG+G+V+KG +G+ VA+K+L S K + F++EV
Sbjct: 299 PIRYSYKEVKKMAGGFKDKLGEGGYGSVFKGKLGSGSCVAIKMLGKS--KGNGQDFISEV 472
Query: 66 GTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA--LGYEKLHEIAIG 123
TIGR +H N+V+L GFC + ALVYE+M NGSLD+++ + + L Y++++ I+I
Sbjct: 473 ATIGRTYHQNIVQLIGFCVHGSKRALVYEFMPNGSLDKFIFSKDESIHLSYDRIYNISIE 652
Query: 124 TARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGT 183
ARGIAYLH CE +I+H+DIKP NILLD NF PKV+DFGLAKL +N+ + T RGT
Sbjct: 653 VARGIAYLHYGCEMQILHFDIKPHNILLDENFTPKVSDFGLAKLYPIDNSIVPRTAARGT 832
Query: 184 PGYAAPELWMP--FPITHKCDVYSFGMLLFEIVGRRRNLD-IKNTESQEWFPIWVWKKFD 240
GY APEL+ I+HK DVYS+GMLL E+ +R+NL+ SQ +FP W++
Sbjct: 833 IGYMAPELFYNNIGGISHKADVYSYGMLLMEMASKRKNLNPHAERSSQLFFPFWIYNHIG 1012
Query: 241 AGLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLE 294
EE + + + E+ + +M+ VALWC+Q + RP M+ VV+MLEG +E
Sbjct: 1013D---EEDIEMEDVTEEEK----KMIIVALWCIQLKPNDRPSMNKVVEMLEGDIE 1153
>TC207390 weakly similar to UP|Q9ATQ5 (Q9ATQ5) LRK33, partial (34%)
Length = 1342
Score = 240 bits (613), Expect = 6e-64
Identities = 128/308 (41%), Positives = 191/308 (61%), Gaps = 8/308 (2%)
Frame = +3
Query: 4 EKPIRFTSQQLRIATDNYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMA 63
EKP RFT L+ T + LG G G V++G +N +VAVK+L + + ++F+
Sbjct: 198 EKPARFTYADLKRITGGFKEKLGEGAHGAVFRGKLSNEILVAVKILNNTEGEG--KEFIN 371
Query: 64 EVGTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLL---HEKNALGYEKLHEI 120
EVG +G+IHH N+V+L GFC E ALVY NGSL R+++ + + LG+EKL +I
Sbjct: 372 EVGIMGKIHHINVVRLLGFCAEGFHRALVYNLFPNGSLQRFIVPPDDKDHFLGWEKLQQI 551
Query: 121 AIGTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGG 180
A+G A+GI YLHE C I+H+DI P N+LLD NF PK++DFGLAKLC++ + ++MT
Sbjct: 552 ALGIAKGIEYLHEGCNQPIIHFDINPHNVLLDDNFTPKISDFGLAKLCSKNPSLVSMTAA 731
Query: 181 RGTPGYAAPELWMP--FPITHKCDVYSFGMLLFEIVGRRRNLDIKNTES-QEWFPIWVWK 237
RGT GY APE++ +++K D+YS+GMLL E+VG R+N+D+ + + +P W+
Sbjct: 732 RGTLGYIAPEVFSKNFGNVSYKSDIYSYGMLLLEMVGGRKNVDMSSAQDFHVLYPDWIHN 911
Query: 238 KFDAGLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLE--GSLEI 295
D + C I +IA+++ V LWC+Q++ RP + V++MLE G ++
Sbjct: 912 LIDGDVHIHVEDECDI-----KIAKKLAIVGLWCIQWQPVNRPSIKSVIQMLETGGESQL 1076
Query: 296 PKTFNPFQ 303
NPFQ
Sbjct: 1077NVPPNPFQ 1100
>TC220351 similar to UP|Q961E7 (Q961E7) GH28523p (CG1830-PB) , partial (11%)
Length = 1064
Score = 217 bits (552), Expect = 7e-57
Identities = 123/260 (47%), Positives = 159/260 (60%), Gaps = 9/260 (3%)
Frame = +1
Query: 44 VAVKVLRGSSDKKIEEQFMAEVGTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDR 103
VAVK L G E+QF EV TI HH NLV+L GFC E LVYE+M NGSLD
Sbjct: 1 VAVKQLEGIEQG--EKQFRMEVATISSTHHLNLVRLIGFCSEGRHRLLVYEFMKNGSLDD 174
Query: 104 YLL----HEKNALGYEKLHEIAIGTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKV 159
+L H L +E IA+GTARGI YLHE C IVH DIKP NILLD N+ KV
Sbjct: 175 FLFLTEQHSGKLLNWEYRFNIALGTARGITYLHEECRDCIVHCDIKPENILLDENYVAKV 354
Query: 160 ADFGLAKLCN-RENTHITMTGGRGTPGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRR 218
+DFGLAKL N +++ H T+T RGT GY APE PIT K DVY +GM+L EIV RR
Sbjct: 355 SDFGLAKLINPKDHRHRTLTSVRGTRGYLAPEWLANLPITSKSDVYGYGMVLLEIVSGRR 534
Query: 219 NLDIKNTESQEWFPIWVWKKFD----AGLLEEAMIVCGIEEKNREIAERMVKVALWCVQY 274
N D+ +++ F IW +++F+ +G+L++ + +E + E R ++ + WC+Q
Sbjct: 535 NFDVSEETNRKKFSIWAYEEFEKGNISGILDKRL---ANQEVDMEQVRRAIQASFWCIQE 705
Query: 275 RQQLRPMMSDVVKMLEGSLE 294
+ RP MS V++MLEG E
Sbjct: 706 QPSHRPTMSRVLQMLEGVTE 765
>TC231087 similar to UP|Q39203 (Q39203) Receptor-like protein kinase
precursor, partial (30%)
Length = 826
Score = 213 bits (541), Expect = 1e-55
Identities = 126/278 (45%), Positives = 167/278 (59%), Gaps = 14/278 (5%)
Frame = +1
Query: 25 LGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGTIGRIHHFNLVKLFGFCF 84
+G GGFGTV++G ++ ++VAVK L E++F AEV TIG I H NLV+L GFC
Sbjct: 1 VGHGGFGTVFQGELSDASVVAVKRLERPGGG--EKEFRAEVSTIGNIQHVNLVRLRGFCS 174
Query: 85 EKNLIALVYEYMGNGSLDRYLLHEKNALGYEKLHEIAIGTARGIAYLHELCEHRIVHYDI 144
E + LVYEYM NG+L+ YL E L ++ +A+GTA+GIAYLHE C I+H DI
Sbjct: 175 ENSHRLLVYEYMQNGALNVYLRKEGPCLSWDVRFRVAVGTAKGIAYLHEECRCCIIHCDI 354
Query: 145 KPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPGYAAPELWMPFPITHKCDVY 204
KP NILLDG+F KV+DFGLAKL R+ + + +T RGT GY APE IT K DVY
Sbjct: 355 KPENILLDGDFTAKVSDFGLAKLIGRDFSRVLVT-MRGTWGYVAPEWISGVAITTKADVY 531
Query: 205 SFGMLLFEIVGRRRNLDIK--------NTESQE------WFPIWVWKKFDAGLLEEAMIV 250
S+GM L E++G RRN++ ES + +FP W ++ G + + M
Sbjct: 532 SYGMTLLELIGGRRNVEAPLSAGGGGGGGESGDEMGGKWFFPPWAAQRIIEGNVSDVMDK 711
Query: 251 CGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKM 288
N E A R+ VA+WC+Q + +RP M VVKM
Sbjct: 712 RLGNAYNIEEARRVALVAVWCIQDDEAMRPTMGMVVKM 825
>TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial (28%)
Length = 1358
Score = 210 bits (535), Expect = 7e-55
Identities = 133/325 (40%), Positives = 187/325 (56%), Gaps = 17/325 (5%)
Frame = +1
Query: 22 SNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGTIGRIHHFNLVKLFG 81
+N +G GGFG VYKG F++GT++AVK L S ++ +F+ E+G I + H +LVKL+G
Sbjct: 4 ANKIGEGGFGPVYKGCFSDGTLIAVKQL-SSKSRQGNREFLNEIGMISALQHPHLVKLYG 180
Query: 82 FCFEKNLIALVYEYMGNGSLDRYLLHEKN---ALGYEKLHEIAIGTARGIAYLHELCEHR 138
C E + + LVYEYM N SL R L + L + ++I +G ARG+AYLHE +
Sbjct: 181 CCVEGDQLLLVYEYMENNSLARALFGAEEHQIKLDWTTRYKICVGIARGLAYLHEESRLK 360
Query: 139 IVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPGYAAPELWMPFPIT 198
IVH DIK N+LLD + NPK++DFGLAKL +NTHI+ T GT GY APE M +T
Sbjct: 361 IVHRDIKATNVLLDQDLNPKISDFGLAKLDEEDNTHIS-TRIAGTFGYMAPEYAMHGYLT 537
Query: 199 HKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVWKKFDAGLLEEAMIVCGIEEK-- 256
K DVYSFG++ EI+ R N + + +E F + W A LL E + + ++
Sbjct: 538 DKADVYSFGIVALEIINGRSN--TIHRQKEESFSVLEW----AHLLREKGDIMDLVDRRL 699
Query: 257 ----NREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLEIPKTFN-PFQHLIDETK- 310
N+E A M+KVAL C LRP MS VV MLEG + + + F+ ++DE K
Sbjct: 700 GLEFNKEEALVMIKVALLCTNVTAALRPTMSSVVSMLEGKIVVDEEFSGETTEVLDEKKM 879
Query: 311 ------FTTHSDQESNTYTTSVSSV 329
+ S+ + +T S +SV
Sbjct: 880 EKMRLYYQELSNSKEEPWTASSTSV 954
>TC220244 weakly similar to UP|Q9LL58 (Q9LL58) Receptor-like kinase, partial
(32%)
Length = 1112
Score = 204 bits (520), Expect = 4e-53
Identities = 114/257 (44%), Positives = 166/257 (64%), Gaps = 8/257 (3%)
Frame = +1
Query: 43 MVAVKVLRGS-SDKKIEEQFMAEVGTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSL 101
+VAVK+L + D K F+ EVGT+G+IHH N+V+L GFC + ALVY++ NGSL
Sbjct: 37 LVAVKILNDTVGDGK---DFINEVGTMGKIHHVNVVRLLGFCADGFHRALVYDFFPNGSL 207
Query: 102 DRYLLHEKNA---LGYEKLHEIAIGTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPK 158
R+L N LG+EKL +IA+G A+G+ YLH C+ RI+H+DI P NIL+D +F PK
Sbjct: 208 QRFLAPPDNKDVFLGWEKLQQIALGVAKGVEYLHLGCDQRILHFDINPHNILIDDHFVPK 387
Query: 159 VADFGLAKLCNRENTHITMTGGRGTPGYAAPELWMP--FPITHKCDVYSFGMLLFEIVGR 216
+ DFGLAKLC + + +++T RGT GY APE++ +++K D+YS+GMLL E+VG
Sbjct: 388 ITDFGLAKLCPKNQSTVSITAARGTLGYIAPEVFSRNFGNVSYKSDIYSYGMLLLEMVGG 567
Query: 217 RRNLDIKNTES-QEWFPIWVWKKFDAGLLEEAMIVCGIE-EKNREIAERMVKVALWCVQY 274
R+N ++ ES Q +P W+ LL+ + IE E + IA+++ V LWC+++
Sbjct: 568 RKNTNMSAEESFQVLYPEWI-----HNLLKSRDVQVTIEDEGDVRIAKKLAIVGLWCIEW 732
Query: 275 RQQLRPMMSDVVKMLEG 291
RP M V++MLEG
Sbjct: 733 NPIDRPSMKTVIQMLEG 783
>TC217441 homologue to GB|AAK68074.1|14573459|AF384970 somatic embryogenesis
receptor-like kinase 3 {Arabidopsis thaliana;} , partial
(54%)
Length = 1008
Score = 200 bits (509), Expect = 7e-52
Identities = 119/294 (40%), Positives = 174/294 (58%), Gaps = 10/294 (3%)
Frame = +2
Query: 8 RFTSQQLRIATDNYSN--LLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEV 65
RF+ ++L++ATDN+SN +LG GGFG VYKG +G++VAVK L+ + QF EV
Sbjct: 116 RFSLRELQVATDNFSNKHILGRGGFGKVYKGRLADGSLVAVKRLKEERTQGG*LQFQTEV 295
Query: 66 GTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA---LGYEKLHEIAI 122
I H NL++L GFC LVY YM NGS+ L + + LG+ + IA+
Sbjct: 296 EMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERQESQPPLGWPERKRIAL 475
Query: 123 GTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRG 182
G+ARG+AYLH+ C+ +I+H D+K NILLD F V DFGLAKL + ++TH+T T RG
Sbjct: 476 GSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVT-TAVRG 652
Query: 183 TPGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVWKKFDAG 242
T G+ APE + K DV+ +G++L E++ +R D+ + + + W K G
Sbjct: 653 TIGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVK---G 823
Query: 243 LLEEAMIVCGIE-----EKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEG 291
LL++ + ++ N E E++++VAL C Q RP MS+VV+MLEG
Sbjct: 824 LLKDRKLETLVDADLQGSYNDEEVEQLIQVALLCTQGSPMERPKMSEVVRMLEG 985
>TC209519 similar to UP|Q84K89 (Q84K89) Receptor kinase LRK10 (Receptor
kinase LRK14), partial (19%)
Length = 866
Score = 195 bits (495), Expect = 3e-50
Identities = 120/278 (43%), Positives = 171/278 (61%), Gaps = 11/278 (3%)
Frame = +2
Query: 83 CFEKNLIALVYEYMGNGSLDRYLL--HEKNALGYEKLHEIAIGTARGIAYLHELCEHRIV 140
C E ALVYE+M NGSLD+Y+ E +L Y+K +EI +G ARGIAYLH+ C+ +I+
Sbjct: 2 CAEGEKHALVYEFMPNGSLDKYIFSKEESVSLSYDKTYEICLGIARGIAYLHQDCDVQIL 181
Query: 141 HYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPGYAAPELWMP--FPIT 198
H+DIKP NILLD NF PKV+DFGLAKL ++ I +TG RGT GY APEL+ ++
Sbjct: 182 HFDIKPHNILLDDNFIPKVSDFGLAKLYPIKDKSIILTGLRGTFGYMAPELFYKNIGGVS 361
Query: 199 HKCDVYSFGMLLFEIVGRRRNLDIKNTE--SQEWFPIWVWKKFDAGLLEEAMIVCGIEEK 256
+K DVYSFGMLL E+ RRRN + +TE SQ +FP W++ F + E+ + + + E+
Sbjct: 362 YKADVYSFGMLLMEMGSRRRNSN-PHTEHSSQHFFPFWIYDHF---MEEKDIHMEEVSEE 529
Query: 257 NREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLE----IPK-TFNPFQHLIDETKF 311
++ + ++M V+LWC+Q + RP M VV+MLEG +E PK F P + ID +
Sbjct: 530 DKILVKKMFIVSLWCIQLKPNDRPSMKKVVEMLEGKVENIDMPPKPVFYPHETTIDSDQ- 706
Query: 312 TTHSDQESNTYTTSVSSVMVSDSNIVCATPIMRKYEIE 349
+ SD S+ + S N T +M K+ ++
Sbjct: 707 ASWSDSTSSCKNIDKTKSNFSLENNS*QTMVMNKFNLQ 820
>TC218138 homologue to UP|Q8GRK2 (Q8GRK2) Somatic embryogenesis receptor
kinase 1, partial (60%)
Length = 1444
Score = 192 bits (487), Expect = 3e-49
Identities = 118/294 (40%), Positives = 171/294 (58%), Gaps = 10/294 (3%)
Frame = +1
Query: 8 RFTSQQLRIATDNYSN--LLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEV 65
RF+ ++L++ATD++SN +LG GGFG VYKG +G++VAVK L+ E QF EV
Sbjct: 118 RFSLRELQVATDSFSNKNILGRGGFGKVYKGRLADGSLVAVKRLKEERTPGGELQFQTEV 297
Query: 66 GTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHE---KNALGYEKLHEIAI 122
I H NL++L GFC LVY YM NGS+ L + L + +A+
Sbjct: 298 EMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPPYQEPLDWPTRKRVAL 477
Query: 123 GTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRG 182
G+ARG++YLH+ C+ +I+H D+K NILLD F V DFGLAKL + ++TH+T T RG
Sbjct: 478 GSARGLSYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVT-TAVRG 654
Query: 183 TPGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVWKKFDAG 242
T G+ APE + K DV+ +G++L E++ +R D+ + + + W K G
Sbjct: 655 TIGHIAPEYLSTGKSSEKTDVFGYGIMLLELITGQRAFDLARLANDDDVMLLDWVK---G 825
Query: 243 LLEE---AMIVCGIEEKN--REIAERMVKVALWCVQYRQQLRPMMSDVVKMLEG 291
LL+E M+V + N E++++VAL C Q RP MS+VV+MLEG
Sbjct: 826 LLKEKKLEMLVDPDLQTNYIETEVEQLIQVALLCTQGSPMDRPKMSEVVRMLEG 987
>TC207391 similar to UP|Q9SWU8 (Q9SWU8) Receptor-like kinase (Fragment),
partial (29%)
Length = 963
Score = 149 bits (375), Expect(2) = 1e-48
Identities = 69/136 (50%), Positives = 97/136 (70%), Gaps = 3/136 (2%)
Frame = +1
Query: 60 QFMAEVGTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLL---HEKNALGYEK 116
+F+ EV +G+IHH N+V+L G+C E ALVY + NGSL ++ ++N LG+EK
Sbjct: 16 RFINEVEIMGKIHHINVVRLLGYCAEGIHRALVYNFFPNGSLQSFIFPPDDKQNFLGWEK 195
Query: 117 LHEIAIGTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHIT 176
L IA+G A+GI YLH+ C H I+H+DI P N+LLD NF PK++DFGLAKLC++ + ++
Sbjct: 196 LQNIALGIAKGIGYLHQGCNHPIIHFDINPHNVLLDDNFTPKISDFGLAKLCSKNPSLVS 375
Query: 177 MTGGRGTPGYAAPELW 192
MT RGT GY APE++
Sbjct: 376 MTAARGTLGYIAPEVF 423
Score = 62.8 bits (151), Expect(2) = 1e-48
Identities = 37/109 (33%), Positives = 62/109 (55%), Gaps = 3/109 (2%)
Frame = +2
Query: 197 ITHKCDVYSFGMLLFEIVGRRRNLDIKNTES-QEWFPIWVWKKFDAGLLEEAMIVCGIEE 255
+++K D+YS+GMLL E+VG R+N+D + E +P W+ L+ + + +E
Sbjct: 443 VSYKSDIYSYGMLLLEMVGGRKNVDTSSPEDFHVLYPDWM-----HDLVHGDVHIHVEDE 607
Query: 256 KNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLEIPKTF--NPF 302
+ +IA ++ V LWC+Q++ RP + V++MLE E T NPF
Sbjct: 608 GDVKIARKLAIVGLWCIQWQPLNRPSIKSVIQMLESKEEDLLTVPPNPF 754
>TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-like protein,
partial (54%)
Length = 2032
Score = 187 bits (476), Expect = 5e-48
Identities = 121/319 (37%), Positives = 171/319 (52%), Gaps = 5/319 (1%)
Frame = +3
Query: 7 IRFTSQQLRIATDNYS--NLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAE 64
++F + AT+ +S N LG GGFG VYKG ++G +VAVK L SS + EE F E
Sbjct: 786 LQFDFSTIEAATNKFSADNKLGEGGFGEVYKGTLSSGQVVAVKRLSKSSGQGGEE-FKNE 962
Query: 65 VGTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLL--HEKNALGYEKLHEIAI 122
V + ++ H NLV+L GFC + LVYEY+ N SLD L ++ L + + ++I
Sbjct: 963 VVVVAKLQHRNLVRLLGFCLQGEEKILVYEYVPNKSLDYILFDPEKQRELDWGRRYKIIG 1142
Query: 123 GTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRG 182
G ARGI YLHE RI+H D+K NILLDG+ NPK++DFG+A++ + T + G
Sbjct: 1143 GIARGIQYLHEDSRLRIIHRDLKASNILLDGDMNPKISDFGMARIFGVDQTQGNTSRIVG 1322
Query: 183 TPGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVWKKFDAG 242
T GY APE M + K DVYSFG+LL EI+ ++N T+ E + W+ + G
Sbjct: 1323 TYGYMAPEYAMHGEFSVKSDVYSFGVLLMEILSGKKNSSFYQTDGAEDLLSYAWQLWKDG 1502
Query: 243 LLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLE-GSLEIPKTFNP 301
E M E N+ R + + L CVQ RP M+ +V ML+ ++ +P P
Sbjct: 1503 TPLELMDPILRESYNQNEVIRSIHIGLLCVQEDPADRPTMATIVLMLDSNTVTLPTPTQP 1682
Query: 302 FQHLIDETKFTTHSDQESN 320
F HS + N
Sbjct: 1683 --------AFFVHSGTDPN 1715
>TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-like protein
, partial (47%)
Length = 1305
Score = 187 bits (475), Expect = 6e-48
Identities = 122/337 (36%), Positives = 178/337 (52%), Gaps = 7/337 (2%)
Frame = +2
Query: 7 IRFTSQQLRIATDNYS--NLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAE 64
+RF + AT +S N LG GGFG VYKG+ +G VAVK L S + EE F E
Sbjct: 38 LRFDFSTIEAATQKFSEANKLGEGGFGEVYKGLLPSGQEVAVKRLSKISGQGGEE-FKNE 214
Query: 65 VGTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLL--HEKNALGYEKLHEIAI 122
V + ++ H NLV+L GFC E LVYE++ N SLD L ++ +L + + ++I
Sbjct: 215 VEIVAKLQHRNLVRLLGFCLEGEEKILVYEFVANKSLDYILFDPEKQKSLDWTRRYKIVE 394
Query: 123 GTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRG 182
G ARGI YLHE +I+H D+K N+LLDG+ NPK++DFG+A++ + T G
Sbjct: 395 GIARGIQYLHEDSRLKIIHRDLKASNVLLDGDMNPKISDFGMARIFGVDQTQANTNRIVG 574
Query: 183 TPGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVWKKFDAG 242
T GY +PE M + K DVYSFG+L+ EI+ +RN T+ E + WK +
Sbjct: 575 TYGYMSPEYAMHGEYSAKSDVYSFGVLILEIISGKRNSSFYETDVAEDLLSYAWKLWKDE 754
Query: 243 LLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEG---SLEIPKTF 299
E M E R R + + L CVQ RP M+ VV ML+ +L++P
Sbjct: 755 APLELMDQSLRESYTRNEVIRCIHIGLLCVQEDPIDRPTMASVVLMLDSYSVTLQVPN-- 928
Query: 300 NPFQHLIDETKFTTHSDQESNTYTTSVSSVMVSDSNI 336
P ++ T+ + + TT+ +S V+D ++
Sbjct: 929 QPAFYINSRTEPNMPKGLKIDQSTTNSTSKSVNDMSV 1039
>TC218530 similar to UP|Q9SGU0 (Q9SGU0) T6H22.8.1 protein, partial (25%)
Length = 1190
Score = 183 bits (464), Expect = 1e-46
Identities = 106/252 (42%), Positives = 147/252 (58%)
Frame = +3
Query: 44 VAVKVLRGSSDKKIEEQFMAEVGTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDR 103
+AVK L S + + QF+ E+ TI + H NLVKL+G C E + LVYEY+ N SLD+
Sbjct: 6 IAVKQLSVGSHQG-KSQFITEIATISAVQHRNLVKLYGCCIEGSKRLLVYEYLENKSLDQ 182
Query: 104 YLLHEKNALGYEKLHEIAIGTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFG 163
L + L + ++I +G ARG+ YLHE RIVH D+K NILLD PK++DFG
Sbjct: 183 ALFGKCLTLNWSTRYDICLGVARGLTYLHEESRLRIVHRDVKASNILLDYELIPKISDFG 362
Query: 164 LAKLCNRENTHITMTGGRGTPGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIK 223
LAKL + + THI+ TG GT GY APE M +T K DV+SFG++ E+V R N D
Sbjct: 363 LAKLYDDKKTHIS-TGVAGTIGYLAPEYAMRGHLTEKADVFSFGVVALELVSGRPNSDSS 539
Query: 224 NTESQEWFPIWVWKKFDAGLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMS 283
+ + W W+ + + + ++ + E N E +R+V +AL C Q LRP MS
Sbjct: 540 LEGEKVYLLEWAWQLHEKNCIID-LVDDRLSEFNEEEVKRVVGIALLCTQTSPTLRPSMS 716
Query: 284 DVVKMLEGSLEI 295
VV ML G +E+
Sbjct: 717 RVVAMLSGDIEV 752
>TC231333 similar to UP|Q8L741 (Q8L741) AT3g13690/MMM17_12, partial (38%)
Length = 1009
Score = 179 bits (453), Expect = 2e-45
Identities = 117/304 (38%), Positives = 174/304 (56%), Gaps = 10/304 (3%)
Frame = +3
Query: 9 FTSQQLRIATDNYS--NLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVG 66
FT +L++AT +S N L GGFG+V++G+ +G ++AVK + +S + +++F +EV
Sbjct: 6 FTFSELQLATGGFSQANFLAEGGFGSVHRGVLPDGQVIAVKQYKLASTQG-DKEFCSEVE 182
Query: 67 TIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEK-NALGYEKLHEIAIGTA 125
+ H N+V L GFC + LVYEY+ NGSLD ++ K N L + +IA+G A
Sbjct: 183 VLSCAQHRNVVMLIGFCVDDGRRLLVYEYICNGSLDSHIYRRKQNVLEWSARQKIAVGAA 362
Query: 126 RGIAYLHELCEHR-IVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTP 184
RG+ YLHE C IVH D++P NILL +F V DFGLA+ + + T GT
Sbjct: 363 RGLRYLHEECRVGCIVHRDMRPNNILLTHDFEALVGDFGLARWQPDGDMGVE-TRVIGTF 539
Query: 185 GYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQ----EWFPIWVWKKFD 240
GY APE IT K DVYSFG++L E+V R+ +DI + Q EW + K+
Sbjct: 540 GYLAPEYAQSGQITEKADVYSFGIVLLELVTGRKAVDINRPKGQQCLSEWARPLLEKQAT 719
Query: 241 AGLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLE--IPKT 298
L++ ++ C ++++ RM+K + C+ LRP MS V++MLEG + +P T
Sbjct: 720 YKLIDPSLRNCYVDQE----VYRMLKCSSLCIGRDPHLRPRMSQVLRMLEGDILM*LPIT 887
Query: 299 FNPF 302
F PF
Sbjct: 888 F-PF 896
>TC230360 similar to GB|AAP68229.1|31711746|BT008790 At5g63940 {Arabidopsis
thaliana;} , partial (42%)
Length = 1533
Score = 175 bits (444), Expect = 2e-44
Identities = 97/284 (34%), Positives = 164/284 (57%), Gaps = 5/284 (1%)
Frame = +2
Query: 12 QQLRIATDNYS--NLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGTIG 69
Q+L AT N++ NL+G GG VY+G +G +AVK+L+ S + + ++F+ E+ I
Sbjct: 431 QELLSATSNFASDNLIGRGGCSYVYRGCLPDGKELAVKILKPSEN--VIKEFVQEIEIIT 604
Query: 70 RIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEK---NALGYEKLHEIAIGTAR 126
+ H N++ + GFC E N + LVY+++ GSL+ L K +A G+++ +++A+G A
Sbjct: 605 TLRHKNIISISGFCLEGNHLLLVYDFLSRGSLEENLHGNKVDCSAFGWQERYKVAVGVAE 784
Query: 127 GIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPGY 186
+ YLH C ++H D+K NILL +F P+++DFGLA ++HIT T GT GY
Sbjct: 785 ALDYLHNGCAQAVIHRDVKSSNILLADDFEPQLSDFGLASW-GSSSSHITCTDVAGTFGY 961
Query: 187 AAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVWKKFDAGLLEE 246
APE +M +T K DVY+FG++L E++ R+ ++ ++ + QE +W + G +
Sbjct: 962 LAPEYFMHGRVTDKIDVYAFGVVLLELLSNRKPINNESPKGQESLVMWATPILEGGKFSQ 1141
Query: 247 AMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLE 290
+ E N +RM+ A C++ +LRP ++ + LE
Sbjct: 1142LLDPSLGSEYNTCQIKRMILAATLCIRRIPRLRPQINLALLDLE 1273
>TC216858 homologue to UP|Q76FZ8 (Q76FZ8) Brassinosteroid receptor, partial
(29%)
Length = 1320
Score = 173 bits (438), Expect = 1e-43
Identities = 114/301 (37%), Positives = 170/301 (55%), Gaps = 14/301 (4%)
Frame = +1
Query: 4 EKPIR-FTSQQLRIATDNYSN--LLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQ 60
EKP+R T L AT+ + N L+GSGGFG VYK +G++VA+K L S + + +
Sbjct: 28 EKPLRKLTFADLLDATNGFHNDSLIGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQG-DRE 204
Query: 61 FMAEVGTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA---LGYEKL 117
F AE+ TIG+I H NLV L G+C LVYEYM GSL+ L +K A L +
Sbjct: 205 FTAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDQKKAGIKLNWAIR 384
Query: 118 HEIAIGTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITM 177
+IAIG ARG+A+LH C I+H D+K N+LLD N +V+DFG+A+L + +TH+++
Sbjct: 385 RKIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSV 564
Query: 178 TGGRGTPGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRR---NLDIKNTESQEWF--- 231
+ GTPGY PE + F + K DVYS+G++L E++ +R + D + W
Sbjct: 565 STLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTDSADFGDNNLVGWVKQH 744
Query: 232 -PIWVWKKFDAGLLEEAMIVCGIEEKNREI-AERMVKVALWCVQYRQQLRPMMSDVVKML 289
+ + FD L++ E+ N E+ + +K+A+ C+ R RP M V+ M
Sbjct: 745 AKLKISDIFDPELMK--------EDPNLEMELLQHLKIAVSCLDDRPWRRPTMIQVMAMF 900
Query: 290 E 290
+
Sbjct: 901 K 903
>TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like protein kinase,
RLK3 precursor, partial (35%)
Length = 1304
Score = 172 bits (435), Expect = 3e-43
Identities = 105/323 (32%), Positives = 165/323 (50%), Gaps = 11/323 (3%)
Frame = +3
Query: 20 NYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGTIGRIHHFNLVKL 79
+Y +G GGFG VYKG+ +G +AVK L SS + E F E+ I ++ H NLV L
Sbjct: 228 SYEKRIGEGGFGVVYKGVLPDGREIAVKKLSKSSGQGANE-FKNEILLIAKLQHRNLVTL 404
Query: 80 FGFCFEKNLIALVYEYMGNGSLDRYLL--HEKNALGYEKLHEIAIGTARGIAYLHELCEH 137
GFC E++ L+YE++ N SLD +L H L + + ++I G A+GI+YLHE
Sbjct: 405 LGFCLEEHEKMLIYEFVSNKSLDYFLFDSHRSKQLNWSERYKIIEGIAQGISYLHEHSRL 584
Query: 138 RIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPGYAAPELWMPFPI 197
+++H D+KP N+LLD N NPK++DFG+A++ + GT GY +PE M
Sbjct: 585 KVIHRDLKPSNVLLDSNMNPKISDFGMARIVAIDQLQGKTNRIVGTYGYMSPEYAMHGQF 764
Query: 198 THKCDVYSFGMLLFEIVGRRRNL--------DIKNTESQEWFPIWVWKKFDAGLLEEAMI 249
+ K DV+SFG+++ EI+ +RN D+ + ++W FD + E
Sbjct: 765 SEKSDVFSFGVIVLEIISAKRNTRSVFSDHDDLLSYTWEQWMDEAPLNIFDQSIEAE--- 935
Query: 250 VCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSL-EIPKTFNPFQHLIDE 308
C E + +++ L CVQ + RP ++ V+ L S+ E+P P +
Sbjct: 936 FCDHSE-----VVKCIQIGLLCVQEKPDDRPKITQVISYLNSSITELPLPKKPIRQSGIV 1100
Query: 309 TKFTTHSDQESNTYTTSVSSVMV 331
K +T + + SV +
Sbjct: 1101QKIAVGESSSGSTPSINEMSVSI 1169
>TC229775 weakly similar to UP|KPEL_DROME (Q05652) Probable
serine/threonine-protein kinase pelle , partial (8%)
Length = 1250
Score = 171 bits (433), Expect = 5e-43
Identities = 110/292 (37%), Positives = 160/292 (54%), Gaps = 3/292 (1%)
Frame = +1
Query: 5 KPIRFTSQQLRIATDNYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAE 64
K ++T +L T+N++ +LG GGFG VY G F + T VAVK+L S+ + E QF+AE
Sbjct: 37 KQRQYTFNELVKITNNFTRILGRGGFGKVYHG-FIDDTQVAVKMLSPSAVRGYE-QFLAE 210
Query: 65 VGTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA---LGYEKLHEIA 121
V + R+HH NL L G+C E+N I L+YEYM NG+LD + + + L +E +IA
Sbjct: 211 VKLLMRVHHRNLTSLVGYCNEENNIGLIYEYMANGNLDEIVSGKSSRAKFLTWEDRLQIA 390
Query: 122 IGTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGR 181
+ A+G+ YLH C+ I+H D+K NILL+ NF K+ADFGL+K + T
Sbjct: 391 LDAAQGLEYLHNGCKPPIIHRDVKCANILLNENFQAKLADFGLSKSFPTDGGSYMSTVVA 570
Query: 182 GTPGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVWKKFDA 241
GTPGY PE + +T K DVYSFG++L E+V + I T + WV
Sbjct: 571 GTPGYLDPEYSISSRLTEKSDVYSFGVVLLEMVTGQP--AIAKTPDKTHISQWVKSMLSN 744
Query: 242 GLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSL 293
G ++ E+ + R+V++ + V RP MS +V L+ L
Sbjct: 745 GDIKNIADSRFKEDFDTSSVWRIVEIGMASVSISPFKRPSMSYIVNELKECL 900
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.138 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,371,248
Number of Sequences: 63676
Number of extensions: 219791
Number of successful extensions: 2805
Number of sequences better than 10.0: 944
Number of HSP's better than 10.0 without gapping: 2041
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2067
length of query: 355
length of database: 12,639,632
effective HSP length: 98
effective length of query: 257
effective length of database: 6,399,384
effective search space: 1644641688
effective search space used: 1644641688
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148970.6


