

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148970.5 + phase: 0
(402 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
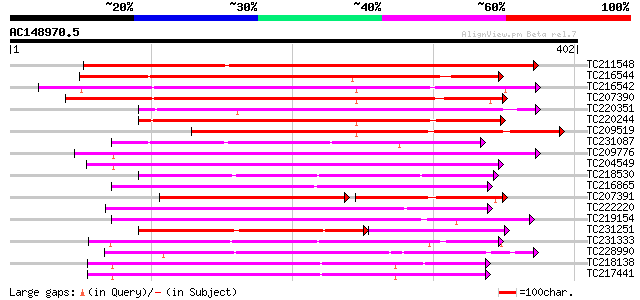
Score E
Sequences producing significant alignments: (bits) Value
TC211548 weakly similar to UP|Q7XIP7 (Q7XIP7) Receptor-like kina... 369 e-102
TC216544 weakly similar to UP|Q9LL55 (Q9LL55) Receptor-like kina... 248 3e-66
TC216542 weakly similar to UP|Q9LL55 (Q9LL55) Receptor-like kina... 248 3e-66
TC207390 weakly similar to UP|Q9ATQ5 (Q9ATQ5) LRK33, partial (34%) 237 8e-63
TC220351 similar to UP|Q961E7 (Q961E7) GH28523p (CG1830-PB) , pa... 220 8e-58
TC220244 weakly similar to UP|Q9LL58 (Q9LL58) Receptor-like kina... 208 3e-54
TC209519 similar to UP|Q84K89 (Q84K89) Receptor kinase LRK10 (Re... 201 4e-52
TC231087 similar to UP|Q39203 (Q39203) Receptor-like protein kin... 199 2e-51
TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-l... 195 3e-50
TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-l... 194 5e-50
TC218530 similar to UP|Q9SGU0 (Q9SGU0) T6H22.8.1 protein, partia... 191 5e-49
TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial... 190 9e-49
TC207391 similar to UP|Q9SWU8 (Q9SWU8) Receptor-like kinase (Fra... 134 3e-48
TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like prot... 185 4e-47
TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.1... 184 8e-47
TC231251 weakly similar to UP|Q39202 (Q39202) Receptor-like prot... 139 7e-46
TC231333 similar to UP|Q8L741 (Q8L741) AT3g13690/MMM17_12, parti... 179 2e-45
TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, comp... 176 1e-44
TC218138 homologue to UP|Q8GRK2 (Q8GRK2) Somatic embryogenesis r... 173 1e-43
TC217441 homologue to GB|AAK68074.1|14573459|AF384970 somatic em... 173 1e-43
>TC211548 weakly similar to UP|Q7XIP7 (Q7XIP7) Receptor-like kinase
TAK33-like protein, partial (58%)
Length = 1194
Score = 369 bits (947), Expect = e-102
Identities = 182/324 (56%), Positives = 234/324 (72%), Gaps = 1/324 (0%)
Frame = +2
Query: 53 KPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLN-CLDMGMEEQFKAE 111
KP+RFT ++L T+ YS +LGSG FGVV+KG SNG VAVKVL D ++EQF AE
Sbjct: 2 KPIRFTDQQLRIATDNYSFLLGSGGFGVVYKGSFSNGTIVAVKVLRGSSDKRIDEQFMAE 181
Query: 112 VITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAI 171
V TIG+ +H NLV+LYGFCF R RALVYEY+ NG+L+KY+F F+KLH+IA+
Sbjct: 182 VGTIGKVHHFNLVRLYGFCFERHLRALVYEYMVNGALEKYLF--HENTTLSFEKLHEIAV 355
Query: 172 GTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRG 231
GTA+GIAYLHEEC+ RIIHYDIKP N+LLD PK+ADFGLAKL +R++ T RG
Sbjct: 356 GTARGIAYLHEECQQRIIHYDIKPGNILLDRNFCPKVADFGLAKLCNRDNTHISMTGGRG 535
Query: 232 TRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENN 291
T GYAAPE+W P+PVT+KCDVYSFG+LLFEI+GRRR+ + + ESQ WFP W WE F+
Sbjct: 536 TPGYAAPELWLPFPVTHKCDVYSFGMLLFEIIGRRRNHNINLPESQVWFPMWVWERFDAE 715
Query: 292 ELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSPPFPF 351
+ +++ C IE+++ EIAER++KVAL CVQY P RP+MS VVKMLEG +++ P PF
Sbjct: 716 NVEDLISACGIEDQNREIAERIVKVALSCVQYKPEARPIMSVVVKMLEGSVEVPKPLNPF 895
Query: 352 HNLVPAKENSTQEGSTADSDTTTS 375
+L+ T + ++T TS
Sbjct: 896 QHLIDWTPPPTDPVQASQTNTDTS 967
>TC216544 weakly similar to UP|Q9LL55 (Q9LL55) Receptor-like kinase, partial
(36%)
Length = 1229
Score = 248 bits (634), Expect = 3e-66
Identities = 132/305 (43%), Positives = 194/305 (63%), Gaps = 4/305 (1%)
Frame = +2
Query: 50 NREKPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFK 109
N P+R++ +++ ++ + LG G +G VFKG+L +G VA+K+L G + F
Sbjct: 287 NNLMPIRYSYKEVKKMAGGFKDKLGEGGYGSVFKGKLGSGSCVAIKMLG-KSKGNGQDFI 463
Query: 110 AEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKI 169
+EV TIGRTYH N+V+L GFC H KRALVYE++ NGSLDK+IF + +++ I
Sbjct: 464 SEVATIGRTYHQNIVQLIGFCVHGSKRALVYEFMPNGSLDKFIFSKDESIHLSYDRIYNI 643
Query: 170 AIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHF 229
+I A+GIAYLH C+ +I+H+DIKP N+LLD PK++DFGLAKL +++I T
Sbjct: 644 SIEVARGIAYLHYGCEMQILHFDIKPHNILLDENFTPKVSDFGLAKLYPIDNSIVPRTAA 823
Query: 230 RGTRGYAAPEMW--KPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQ-WFPRWTWE 286
RGT GY APE++ +++K DVYS+G+LL E+ +R++ + S Q +FP W +
Sbjct: 824 RGTIGYMAPELFYNNIGGISHKADVYSYGMLLMEMASKRKNLNPHAERSSQLFFPFWIYN 1003
Query: 287 MFENNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEID-IS 345
+ E + M + E E+K M+ VALWC+Q PNDRP M+ VV+MLEG+I+ +
Sbjct: 1004HIGDEEDIEMEDVTEEEKK-------MIIVALWCIQLKPNDRPSMNKVVEMLEGDIENLE 1162
Query: 346 SPPFP 350
PP P
Sbjct: 1163IPPKP 1177
>TC216542 weakly similar to UP|Q9LL55 (Q9LL55) Receptor-like kinase, partial
(38%)
Length = 1435
Score = 248 bits (634), Expect = 3e-66
Identities = 137/364 (37%), Positives = 219/364 (59%), Gaps = 8/364 (2%)
Frame = +2
Query: 21 FSSTNNIEVVVQTDSKHEFATMERIFSNI--NREKPVRFTPEKLDEITEKYSTILGSGAF 78
F T I +++ K + E I + + N P+ ++ +++ ++ + LG G +
Sbjct: 263 FGMTLFIVLLIYKWRKRHLSIYENIENYLEQNNLMPIGYSYKEIKKMARGFKEKLGGGGY 442
Query: 79 GVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRAL 138
G+VFKG+L +G +VA+K+L+ G + F +E+ TIGR +H N+V+L G+C KRAL
Sbjct: 443 GIVFKGKLHSGPSVAIKMLHKAK-GNGQDFISEIATIGRIHHQNVVQLIGYCVEGSKRAL 619
Query: 139 VYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENV 198
VYE++ NGSLDK+IF + +++ IAIG A+GIAYLH C+ +I+H+DIKP N+
Sbjct: 620 VYEFMPNGSLDKFIFTKDGNIHLTYDEIYNIAIGVARGIAYLHHGCEMQILHFDIKPHNI 799
Query: 199 LLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPY--PVTYKCDVYSFG 256
LLD PK++DFGLAKL +++I T RGT GY APE++ +++K DVYSFG
Sbjct: 800 LLDETFTPKVSDFGLAKLYPIDNSIVTMTAARGTIGYMAPELFYKNIGGISHKADVYSFG 979
Query: 257 ILLFEIVGRRRHFDSSYSESQQ-WFPRWTWEMFENNELVVMLALCEIEEKDSEIAERMLK 315
+LL E+ +R++ + S Q +FP W + + M + E++++IA++M+
Sbjct: 980 MLLMEMASKRKNLNPHADHSSQLYFPFWIYNQLGKETDIEMEG---VTEEENKIAKKMII 1150
Query: 316 VALWCVQYSPNDRPLMSTVVKMLEGEID-ISSPPFP--FHNLVPAKENSTQEGSTADSDT 372
V+LWC+Q P DRP M+ VV+MLEG+I+ + PP P + + + S T +D
Sbjct: 1151VSLWCIQLKPTDRPSMNKVVEMLEGDIESLEIPPKPSLYPHETMENDQSIYSSQTMSTDF 1330
Query: 373 TTSS 376
+SS
Sbjct: 1331ISSS 1342
>TC207390 weakly similar to UP|Q9ATQ5 (Q9ATQ5) LRK33, partial (34%)
Length = 1342
Score = 237 bits (604), Expect = 8e-63
Identities = 136/320 (42%), Positives = 196/320 (60%), Gaps = 6/320 (1%)
Frame = +3
Query: 40 ATMERIFSNINREKPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNC 99
A + + + EKP RFT L IT + LG GA G VF+G+LSN VAVK+LN
Sbjct: 162 ARVAKFLEDYRAEKPARFTYADLKRITGGFKEKLGEGAHGAVFRGKLSNEILVAVKILNN 341
Query: 100 LDMGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRN 159
+ G ++F EV +G+ +HIN+V+L GFC RALVY NGSL ++I +++
Sbjct: 342 TE-GEGKEFINEVGIMGKIHHINVVRLLGFCAEGFHRALVYNLFPNGSLQRFIVPPDDKD 518
Query: 160 DF-DFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRS 218
F ++KL +IA+G AKGI YLHE C IIH+DI P NVLLD PKI+DFGLAKL S
Sbjct: 519 HFLGWEKLQQIALGIAKGIEYLHEGCNQPIIHFDINPHNVLLDDNFTPKISDFGLAKLCS 698
Query: 219 RESNIELNTHFRGTRGYAAPEMWKPY--PVTYKCDVYSFGILLFEIVGRRRHFD-SSYSE 275
+ ++ T RGT GY APE++ V+YK D+YS+G+LL E+VG R++ D SS +
Sbjct: 699 KNPSLVSMTAARGTLGYIAPEVFSKNFGNVSYKSDIYSYGMLLLEMVGGRKNVDMSSAQD 878
Query: 276 SQQWFPRWTWEMFENNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVV 335
+P W + + + + + C+I +IA+++ V LWC+Q+ P +RP + +V+
Sbjct: 879 FHVLYPDWIHNLIDGDVHIHVEDECDI-----KIAKKLAIVGLWCIQWQPVNRPSIKSVI 1043
Query: 336 KMLE--GEIDISSPPFPFHN 353
+MLE GE ++ PP PF +
Sbjct: 1044QMLETGGESQLNVPPNPFQS 1103
>TC220351 similar to UP|Q961E7 (Q961E7) GH28523p (CG1830-PB) , partial (11%)
Length = 1064
Score = 220 bits (561), Expect = 8e-58
Identities = 123/289 (42%), Positives = 173/289 (59%), Gaps = 4/289 (1%)
Frame = +1
Query: 92 VAVKVLNCLDMGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKY 151
VAVK L ++ G E+QF+ EV TI T+H+NLV+L GFC R LVYE+++NGSLD +
Sbjct: 1 VAVKQLEGIEQG-EKQFRMEVATISSTHHLNLVRLIGFCSEGRHRLLVYEFMKNGSLDDF 177
Query: 152 IFGSKNRND--FDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIA 209
+F ++ + +++ IA+GTA+GI YLHEEC+ I+H DIKPEN+LLD K++
Sbjct: 178 LFLTEQHSGKLLNWEYRFNIALGTARGITYLHEECRDCIVHCDIKPENILLDENYVAKVS 357
Query: 210 DFGLAKL-RSRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRH 268
DFGLAKL ++ T RGTRGY APE P+T K DVY +G++L EIV RR+
Sbjct: 358 DFGLAKLINPKDHRHRTLTSVRGTRGYLAPEWLANLPITSKSDVYGYGMVLLEIVSGRRN 537
Query: 269 FDSSYSESQQWFPRWTWEMFENNELVVML-ALCEIEEKDSEIAERMLKVALWCVQYSPND 327
FD S +++ F W +E FE + +L +E D E R ++ + WC+Q P+
Sbjct: 538 FDVSEETNRKKFSIWAYEEFEKGNISGILDKRLANQEVDMEQVRRAIQASFWCIQEQPSH 717
Query: 328 RPLMSTVVKMLEGEIDISSPPFPFHNLVPAKENSTQEGSTADSDTTTSS 376
RP MS V++MLEG + PP P S EG+ + + T SS
Sbjct: 718 RPTMSRVLQMLEGVTEPERPPAP---------KSVMEGAVSGTSTYLSS 837
>TC220244 weakly similar to UP|Q9LL58 (Q9LL58) Receptor-like kinase, partial
(32%)
Length = 1112
Score = 208 bits (530), Expect = 3e-54
Identities = 114/265 (43%), Positives = 171/265 (64%), Gaps = 5/265 (1%)
Frame = +1
Query: 92 VAVKVLNCLDMGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKY 151
VAVK+LN +G + F EV T+G+ +H+N+V+L GFC RALVY++ NGSL ++
Sbjct: 40 VAVKILNDT-VGDGKDFINEVGTMGKIHHVNVVRLLGFCADGFHRALVYDFFPNGSLQRF 216
Query: 152 IFGSKNRNDF-DFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIAD 210
+ N++ F ++KL +IA+G AKG+ YLH C RI+H+DI P N+L+D PKI D
Sbjct: 217 LAPPDNKDVFLGWEKLQQIALGVAKGVEYLHLGCDQRILHFDINPHNILIDDHFVPKITD 396
Query: 211 FGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPY--PVTYKCDVYSFGILLFEIVGRRRH 268
FGLAKL + + T RGT GY APE++ V+YK D+YS+G+LL E+VG R++
Sbjct: 397 FGLAKLCPKNQSTVSITAARGTLGYIAPEVFSRNFGNVSYKSDIYSYGMLLLEMVGGRKN 576
Query: 269 FDSSYSES-QQWFPRWTWEMFENNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPND 327
+ S ES Q +P W + ++ ++ V + +E D IA+++ V LWC++++P D
Sbjct: 577 TNMSAEESFQVLYPEWIHNLLKSRDVQVTIE----DEGDVRIAKKLAIVGLWCIEWNPID 744
Query: 328 RPLMSTVVKMLEGEID-ISSPPFPF 351
RP M TV++MLEG+ D + +PP PF
Sbjct: 745 RPSMKTVIQMLEGDGDKLIAPPTPF 819
>TC209519 similar to UP|Q84K89 (Q84K89) Receptor kinase LRK10 (Receptor
kinase LRK14), partial (19%)
Length = 866
Score = 201 bits (512), Expect = 4e-52
Identities = 111/268 (41%), Positives = 162/268 (60%), Gaps = 4/268 (1%)
Frame = +2
Query: 130 CFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGIAYLHEECKHRII 189
C +K ALVYE++ NGSLDKYIF + + K ++I +G A+GIAYLH++C +I+
Sbjct: 2 CAEGEKHALVYEFMPNGSLDKYIFSKEESVSLSYDKTYEICLGIARGIAYLHQDCDVQIL 181
Query: 190 HYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPY--PVT 247
H+DIKP N+LLD PK++DFGLAKL + + T RGT GY APE++ V+
Sbjct: 182 HFDIKPHNILLDDNFIPKVSDFGLAKLYPIKDKSIILTGLRGTFGYMAPELFYKNIGGVS 361
Query: 248 YKCDVYSFGILLFEIVGRRRHFD-SSYSESQQWFPRWTWEMFENNELVVMLALCEIEEKD 306
YK DVYSFG+LL E+ RRR+ + + SQ +FP W ++ F + + M E+ E+D
Sbjct: 362 YKADVYSFGMLLMEMGSRRRNSNPHTEHSSQHFFPFWIYDHFMEEKDIHME---EVSEED 532
Query: 307 SEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEID-ISSPPFPFHNLVPAKENSTQEG 365
+ ++M V+LWC+Q PNDRP M VV+MLEG+++ I PP P V +T +
Sbjct: 533 KILVKKMFIVSLWCIQLKPNDRPSMKKVVEMLEGKVENIDMPPKP----VFYPHETTIDS 700
Query: 366 STADSDTTTSSWRTESSRESGFKTKHNA 393
A +TSS + +S F ++N+
Sbjct: 701 DQASWSDSTSSCKNIDKTKSNFSLENNS 784
>TC231087 similar to UP|Q39203 (Q39203) Receptor-like protein kinase
precursor, partial (30%)
Length = 826
Score = 199 bits (505), Expect = 2e-51
Identities = 119/279 (42%), Positives = 160/279 (56%), Gaps = 14/279 (5%)
Frame = +1
Query: 73 LGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIGRTYHINLVKLYGFCFH 132
+G G FG VF+GELS+ VAVK L G E++F+AEV TIG H+NLV+L GFC
Sbjct: 1 VGHGGFGTVFQGELSDASVVAVKRLE-RPGGGEKEFRAEVSTIGNIQHVNLVRLRGFCSE 177
Query: 133 RDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYD 192
R LVYEY++NG+L+ Y+ K + ++A+GTAKGIAYLHEEC+ IIH D
Sbjct: 178 NSHRLLVYEYMQNGALNVYL--RKEGPCLSWDVRFRVAVGTAKGIAYLHEECRCCIIHCD 351
Query: 193 IKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCDV 252
IKPEN+LLD K++DFGLAKL R+ + L T RGT GY APE +T K DV
Sbjct: 352 IKPENILLDGDFTAKVSDFGLAKLIGRDFSRVLVT-MRGTWGYVAPEWISGVAITTKADV 528
Query: 253 YSFGILLFEIVGRRRHFDSSYSE--------------SQQWFPRWTWEMFENNELVVMLA 298
YS+G+ L E++G RR+ ++ S + +FP W + + ++
Sbjct: 529 YSYGMTLLELIGGRRNVEAPLSAGGGGGGGESGDEMGGKWFFPPWAAQRIIEGNVSDVMD 708
Query: 299 LCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKM 337
+ E A R+ VA+WC+Q RP M VVKM
Sbjct: 709 KRLGNAYNIEEARRVALVAVWCIQDDEAMRPTMGMVVKM 825
>TC209776 similar to UP|O65470 (O65470) Serine/threonine kinase-like protein
, partial (47%)
Length = 1305
Score = 195 bits (495), Expect = 3e-50
Identities = 113/333 (33%), Positives = 181/333 (53%), Gaps = 3/333 (0%)
Frame = +2
Query: 47 SNINREKPVRFTPEKLDEITEKYSTI--LGSGAFGVVFKGELSNGENVAVKVLNCLDMGM 104
+ I+ + +RF ++ T+K+S LG G FG V+KG L +G+ VAVK L+ +
Sbjct: 14 TEISAVESLRFDFSTIEAATQKFSEANKLGEGGFGEVYKGLLPSGQEVAVKRLSKISGQG 193
Query: 105 EEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQ 164
E+FK EV + + H NLV+L GFC +++ LVYE+V N SLD +F + + D+
Sbjct: 194 GEEFKNEVEIVAKLQHRNLVRLLGFCLEGEEKILVYEFVANKSLDYILFDPEKQKSLDWT 373
Query: 165 KLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIE 224
+ +KI G A+GI YLHE+ + +IIH D+K NVLLD + PKI+DFG+A++ +
Sbjct: 374 RRYKIVEGIARGIQYLHEDSRLKIIHRDLKASNVLLDGDMNPKISDFGMARIFGVDQTQA 553
Query: 225 LNTHFRGTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWT 284
GT GY +PE + K DVYSFG+L+ EI+ +R+ ++ + +
Sbjct: 554 NTNRIVGTYGYMSPEYAMHGEYSAKSDVYSFGVLILEIISGKRNSSFYETDVAEDLLSYA 733
Query: 285 WEMFENNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEG-EID 343
W+++++ + ++ E R + + L CVQ P DRP M++VV ML+ +
Sbjct: 734 WKLWKDEAPLELMDQSLRESYTRNEVIRCIHIGLLCVQEDPIDRPTMASVVLMLDSYSVT 913
Query: 344 ISSPPFPFHNLVPAKENSTQEGSTADSDTTTSS 376
+ P P + E + +G D TT S+
Sbjct: 914 LQVPNQPAFYINSRTEPNMPKGLKIDQSTTNST 1012
>TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-like protein,
partial (54%)
Length = 2032
Score = 194 bits (494), Expect = 5e-50
Identities = 108/299 (36%), Positives = 171/299 (57%), Gaps = 3/299 (1%)
Frame = +3
Query: 55 VRFTPEKLDEITEKYSTI--LGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEV 112
++F ++ T K+S LG G FG V+KG LS+G+ VAVK L+ E+FK EV
Sbjct: 786 LQFDFSTIEAATNKFSADNKLGEGGFGEVYKGTLSSGQVVAVKRLSKSSGQGGEEFKNEV 965
Query: 113 ITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIG 172
+ + + H NLV+L GFC +++ LVYEYV N SLD +F + + + D+ + +KI G
Sbjct: 966 VVVAKLQHRNLVRLLGFCLQGEEKILVYEYVPNKSLDYILFDPEKQRELDWGRRYKIIGG 1145
Query: 173 TAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGT 232
A+GI YLHE+ + RIIH D+K N+LLD + PKI+DFG+A++ + + GT
Sbjct: 1146 IARGIQYLHEDSRLRIIHRDLKASNILLDGDMNPKISDFGMARIFGVDQTQGNTSRIVGT 1325
Query: 233 RGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNE 292
GY APE + K DVYSFG+LL EI+ +++ ++ + + W+++++
Sbjct: 1326 YGYMAPEYAMHGEFSVKSDVYSFGVLLMEILSGKKNSSFYQTDGAEDLLSYAWQLWKDGT 1505
Query: 293 LVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGE-IDISSPPFP 350
+ ++ E + R + + L CVQ P DRP M+T+V ML+ + + +P P
Sbjct: 1506 PLELMDPILRESYNQNEVIRSIHIGLLCVQEDPADRPTMATIVLMLDSNTVTLPTPTQP 1682
>TC218530 similar to UP|Q9SGU0 (Q9SGU0) T6H22.8.1 protein, partial (25%)
Length = 1190
Score = 191 bits (485), Expect = 5e-49
Identities = 107/255 (41%), Positives = 151/255 (58%)
Frame = +3
Query: 92 VAVKVLNCLDMGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKY 151
+AVK L+ + QF E+ TI H NLVKLYG C KR LVYEY+EN SLD+
Sbjct: 6 IAVKQLSVGSHQGKSQFITEIATISAVQHRNLVKLYGCCIEGSKRLLVYEYLENKSLDQA 185
Query: 152 IFGSKNRNDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADF 211
+FG ++ + I +G A+G+ YLHEE + RI+H D+K N+LLD +L PKI+DF
Sbjct: 186 LFGKCLT--LNWSTRYDICLGVARGLTYLHEESRLRIVHRDVKASNILLDYELIPKISDF 359
Query: 212 GLAKLRSRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDS 271
GLAKL + ++T GT GY APE +T K DV+SFG++ E+V R + DS
Sbjct: 360 GLAKLYD-DKKTHISTGVAGTIGYLAPEYAMRGHLTEKADVFSFGVVALELVSGRPNSDS 536
Query: 272 SYSESQQWFPRWTWEMFENNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLM 331
S + + W W++ E N ++ L + E + E +R++ +AL C Q SP RP M
Sbjct: 537 SLEGEKVYLLEWAWQLHEKN-CIIDLVDDRLSEFNEEEVKRVVGIALLCTQTSPTLRPSM 713
Query: 332 STVVKMLEGEIDISS 346
S VV ML G+I++S+
Sbjct: 714 SRVVAMLSGDIEVST 758
>TC216865 similar to UP|Q9LPG0 (Q9LPG0) T3F20.24 protein, partial (28%)
Length = 1358
Score = 190 bits (483), Expect = 9e-49
Identities = 114/271 (42%), Positives = 154/271 (56%), Gaps = 1/271 (0%)
Frame = +1
Query: 73 LGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIGRTYHINLVKLYGFCFH 132
+G G FG V+KG S+G +AVK L+ +F E+ I H +LVKLYG C
Sbjct: 13 IGEGGFGPVYKGCFSDGTLIAVKQLSSKSRQGNREFLNEIGMISALQHPHLVKLYGCCVE 192
Query: 133 RDKRALVYEYVENGSLDKYIFGSKNRN-DFDFQKLHKIAIGTAKGIAYLHEECKHRIIHY 191
D+ LVYEY+EN SL + +FG++ D+ +KI +G A+G+AYLHEE + +I+H
Sbjct: 193 GDQLLLVYEYMENNSLARALFGAEEHQIKLDWTTRYKICVGIARGLAYLHEESRLKIVHR 372
Query: 192 DIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCD 251
DIK NVLLD L PKI+DFGLAKL E N ++T GT GY APE +T K D
Sbjct: 373 DIKATNVLLDQDLNPKISDFGLAKL-DEEDNTHISTRIAGTFGYMAPEYAMHGYLTDKAD 549
Query: 252 VYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNELVVMLALCEIEEKDSEIAE 311
VYSFGI+ EI+ R + E W + E +++ ++ E + E A
Sbjct: 550 VYSFGIVALEIINGRSNTIHRQKEESFSVLEWAHLLREKGDIMDLVDRRLGLEFNKEEAL 729
Query: 312 RMLKVALWCVQYSPNDRPLMSTVVKMLEGEI 342
M+KVAL C + RP MS+VV MLEG+I
Sbjct: 730 VMIKVALLCTNVTAALRPTMSSVVSMLEGKI 822
>TC207391 similar to UP|Q9SWU8 (Q9SWU8) Receptor-like kinase (Fragment),
partial (29%)
Length = 963
Score = 134 bits (338), Expect(2) = 3e-48
Identities = 68/136 (50%), Positives = 91/136 (66%), Gaps = 1/136 (0%)
Frame = +1
Query: 107 QFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDF-DFQK 165
+F EV +G+ +HIN+V+L G+C RALVY + NGSL +IF ++ +F ++K
Sbjct: 16 RFINEVEIMGKIHHINVVRLLGYCAEGIHRALVYNFFPNGSLQSFIFPPDDKQNFLGWEK 195
Query: 166 LHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIEL 225
L IA+G AKGI YLH+ C H IIH+DI P NVLLD PKI+DFGLAKL S+ ++
Sbjct: 196 LQNIALGIAKGIGYLHQGCNHPIIHFDINPHNVLLDDNFTPKISDFGLAKLCSKNPSLVS 375
Query: 226 NTHFRGTRGYAAPEMW 241
T RGT GY APE++
Sbjct: 376 MTAARGTLGYIAPEVF 423
Score = 75.9 bits (185), Expect(2) = 3e-48
Identities = 39/111 (35%), Positives = 69/111 (62%), Gaps = 3/111 (2%)
Frame = +2
Query: 246 VTYKCDVYSFGILLFEIVGRRRHFDSSYSES-QQWFPRWTWEMFENNELVVMLALCEIEE 304
V+YK D+YS+G+LL E+VG R++ D+S E +P W ++ + + + +E
Sbjct: 443 VSYKSDIYSYGMLLLEMVGGRKNVDTSSPEDFHVLYPDWMHDLVHGDVHIHVE-----DE 607
Query: 305 KDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEID--ISSPPFPFHN 353
D +IA ++ V LWC+Q+ P +RP + +V++MLE + + ++ PP PFH+
Sbjct: 608 GDVKIARKLAIVGLWCIQWQPLNRPSIKSVIQMLESKEEDLLTVPPNPFHS 760
>TC222220 weakly similar to UP|Q9ZP16 (Q9ZP16) Receptor-like protein kinase,
RLK3 precursor, partial (35%)
Length = 1304
Score = 185 bits (469), Expect = 4e-47
Identities = 99/275 (36%), Positives = 154/275 (56%), Gaps = 1/275 (0%)
Frame = +3
Query: 69 YSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIGRTYHINLVKLYG 128
Y +G G FGVV+KG L +G +AVK L+ +FK E++ I + H NLV L G
Sbjct: 231 YEKRIGEGGFGVVYKGVLPDGREIAVKKLSKSSGQGANEFKNEILLIAKLQHRNLVTLLG 410
Query: 129 FCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGIAYLHEECKHRI 188
FC ++ L+YE+V N SLD ++F S ++ + +KI G A+GI+YLHE + ++
Sbjct: 411 FCLEEHEKMLIYEFVSNKSLDYFLFDSHRSKQLNWSERYKIIEGIAQGISYLHEHSRLKV 590
Query: 189 IHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPYPVTY 248
IH D+KP NVLLD + PKI+DFG+A++ + + GT GY +PE +
Sbjct: 591 IHRDLKPSNVLLDSNMNPKISDFGMARIVAIDQLQGKTNRIVGTYGYMSPEYAMHGQFSE 770
Query: 249 KCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMF-ENNELVVMLALCEIEEKDS 307
K DV+SFG+++ EI+ +R+ S +S+ +TWE + + L + E E D
Sbjct: 771 KSDVFSFGVIVLEIISAKRNTRSVFSDHDDLL-SYTWEQWMDEAPLNIFDQSIEAEFCDH 947
Query: 308 EIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEI 342
+ +++ L CVQ P+DRP ++ V+ L I
Sbjct: 948 SEVVKCIQIGLLCVQEKPDDRPKITQVISYLNSSI 1052
>TC219154 similar to PIR|T05754|T05754 S-receptor kinase M4I22.110 precursor
- Arabidopsis thaliana {Arabidopsis thaliana;} ,
partial (31%)
Length = 1363
Score = 184 bits (466), Expect = 8e-47
Identities = 115/304 (37%), Positives = 164/304 (53%), Gaps = 4/304 (1%)
Frame = +1
Query: 73 LGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIGRTYHINLVKLYGFCFH 132
+G G FG V+KG+L G+ +AVK L+ L +F EV I + H NLVKL G C
Sbjct: 367 IGEGGFGPVYKGKLVGGQEIAVKRLSSLSGQGITEFITEVKLIAKLQHRNLVKLLGCCIK 546
Query: 133 RDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYD 192
++ LVYEYV NGSL+ +IF D+ + I +G A+G+ YLH++ + RIIH D
Sbjct: 547 GQEKLLVYEYVVNGSLNSFIFDQIKSKLLDWPRRFNIILGIARGLLYLHQDSRLRIIHRD 726
Query: 193 IKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCDV 252
+K NVLLD KL PKI+DFG+A+ + GT GY APE + K DV
Sbjct: 727 LKASNVLLDEKLNPKISDFGMARAFGGDQTEGNTNRVVGTYGYMAPEYAFDGNFSIKSDV 906
Query: 253 YSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNELVVMLALCEIEEKDSEIAER 312
+SFGILL EIV ++ + + W +++ L L + KDS +
Sbjct: 907 FSFGILLLEIVCGIKNKSLCHENQTLNLVGYAWALWKEQN---ALQLIDSGIKDSCVIPE 1077
Query: 313 MLK---VALWCVQYSPNDRPLMSTVVKMLEGEIDISSPPFP-FHNLVPAKENSTQEGSTA 368
+L+ V+L CVQ P DRP M++V++ML E+D+ P P F KE + +E ++
Sbjct: 1078VLRCIHVSLLCVQQYPEDRPTMTSVIQMLGSEMDMVEPKEPGFFPRRILKEGNLKEMTSN 1257
Query: 369 DSDT 372
D T
Sbjct: 1258DELT 1269
>TC231251 weakly similar to UP|Q39202 (Q39202) Receptor-like protein kinase
precursor , partial (27%)
Length = 905
Score = 139 bits (350), Expect(2) = 7e-46
Identities = 77/164 (46%), Positives = 108/164 (64%), Gaps = 1/164 (0%)
Frame = +1
Query: 92 VAVKVLNCLDMG-MEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDK 150
VAVK LN + ++++FK E+ IG T+H NLV+L GFC +D+R LVYEY+ NG+L
Sbjct: 19 VAVKRLNTFLLEEVQKEFKNELNVIGLTHHKNLVRLLGFCETQDERLLVYEYMSNGTLAS 198
Query: 151 YIFGSKNRNDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIAD 210
+F + + ++ +IA G A+G+ YLHEEC +IIH DIKP+N+LLD +I+D
Sbjct: 199 LVFNVEKPS---WKLRLQIATGVARGLLYLHEECSTQIIHCDIKPQNILLDDYYNARISD 369
Query: 211 FGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCDVYS 254
FGLAK+ + + NT RGT+GY A E +K P+T K DVYS
Sbjct: 370 FGLAKILNMNQS-RTNTAIRGTKGYVALEWFKNMPITAKGDVYS 498
Score = 63.2 bits (152), Expect(2) = 7e-46
Identities = 31/100 (31%), Positives = 51/100 (51%)
Frame = +2
Query: 255 FGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNELVVMLALCEIEEKDSEIAERML 314
+G+LL EIV R+ + E + W ++ + L ++ + D + E+++
Sbjct: 500 YGVLLLEIVSCRKSVEFEADEEKAILTEWAFDCYTEGVLHDLVENDKEALDDMKTLEKLV 679
Query: 315 KVALWCVQYSPNDRPLMSTVVKMLEGEIDISSPPFPFHNL 354
+ALWCVQ P RP M V +MLEG +++ PP P L
Sbjct: 680 MIALWCVQEDPGLRPTMRNVTQMLEGVVEVQIPPCPSSQL 799
>TC231333 similar to UP|Q8L741 (Q8L741) AT3g13690/MMM17_12, partial (38%)
Length = 1009
Score = 179 bits (455), Expect = 2e-45
Identities = 113/304 (37%), Positives = 167/304 (54%), Gaps = 10/304 (3%)
Frame = +3
Query: 57 FTPEKLDEITEKYS--TILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVIT 114
FT +L T +S L G FG V +G L +G+ +AVK +++F +EV
Sbjct: 6 FTFSELQLATGGFSQANFLAEGGFGSVHRGVLPDGQVIAVKQYKLASTQGDKEFCSEVEV 185
Query: 115 IGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTA 174
+ H N+V L GFC +R LVYEY+ NGSLD +I+ K +N ++ KIA+G A
Sbjct: 186 LSCAQHRNVVMLIGFCVDDGRRLLVYEYICNGSLDSHIYRRK-QNVLEWSARQKIAVGAA 362
Query: 175 KGIAYLHEECK-HRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTR 233
+G+ YLHEEC+ I+H D++P N+LL E + DFGLA+ + + ++ + T GT
Sbjct: 363 RGLRYLHEECRVGCIVHRDMRPNNILLTHDFEALVGDFGLARWQP-DGDMGVETRVIGTF 539
Query: 234 GYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNEL 293
GY APE + +T K DVYSFGI+L E+V R+ D + + QQ W + E
Sbjct: 540 GYLAPEYAQSGQITEKADVYSFGIVLLELVTGRKAVDINRPKGQQCLSEWARPLLEKQAT 719
Query: 294 VVM----LALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSP-- 347
+ L C ++++ RMLK + C+ P+ RP MS V++MLEG+I + P
Sbjct: 720 YKLIDPSLRNCYVDQE----VYRMLKCSSLCIGRDPHLRPRMSQVLRMLEGDILM*LPIT 887
Query: 348 -PFP 350
PFP
Sbjct: 888 FPFP 899
>TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, complete
Length = 3346
Score = 176 bits (447), Expect = 1e-44
Identities = 109/311 (35%), Positives = 162/311 (52%), Gaps = 3/311 (0%)
Frame = +1
Query: 68 KYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQ--FKAEVITIGRTYHINLVK 125
K I+G G G+V+KG + NG+NVAVK L + G F AE+ T+GR H ++V+
Sbjct: 2158 KEDNIIGKGGAGIVYKGAMPNGDNVAVKRLPAMSRGSSHDHGFNAEIQTLGRIRHRHIVR 2337
Query: 126 LYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGIAYLHEECK 185
L GFC + + LVYEY+ NGSL + + G K + + +KIA+ AKG+ YLH +C
Sbjct: 2338 LLGFCSNHETNLLVYEYMPNGSLGEVLHGKKGGH-LHWDTRYKIAVEAAKGLCYLHHDCS 2514
Query: 186 HRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPYP 245
I+H D+K N+LLD E +ADFGLAK E + G+ GY APE
Sbjct: 2515 PLIVHRDVKSNNILLDSNFEAHVADFGLAKFLQDSGASECMSAIAGSYGYIAPEYAYTLK 2694
Query: 246 VTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNELVVMLAL-CEIEE 304
V K DVYSFG++L E+V R+ + + +W +M ++N+ V+ L +
Sbjct: 2695 VDEKSDVYSFGVVLLELVTGRKPV-GEFGDGVD-IVQWVRKMTDSNKEGVLKVLDSRLPS 2868
Query: 305 KDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSPPFPFHNLVPAKENSTQE 364
+ VA+ CV+ +RP M VV++L ++ PP H + E+S
Sbjct: 2869 VPLHEVMHVFYVAMLCVEEQAVERPTMREVVQIL---TELPKPPSSKHAIT---ESSLSS 3030
Query: 365 GSTADSDTTTS 375
++ S TT S
Sbjct: 3031 SNSLGSPTTAS 3063
Score = 29.3 bits (64), Expect = 3.3
Identities = 13/30 (43%), Positives = 17/30 (56%)
Frame = +3
Query: 216 LRSRESNIELNTHFRGTRGYAAPEMWKPYP 245
L SR SNIE + HF A+P + P+P
Sbjct: 42 LSSRHSNIEFDFHFHSQTENASPRLVLPFP 131
>TC218138 homologue to UP|Q8GRK2 (Q8GRK2) Somatic embryogenesis receptor
kinase 1, partial (60%)
Length = 1444
Score = 173 bits (439), Expect = 1e-43
Identities = 111/293 (37%), Positives = 166/293 (55%), Gaps = 7/293 (2%)
Frame = +1
Query: 56 RFTPEKLDEITEKYST--ILGSGAFGVVFKGELSNGENVAVKVLNC-LDMGMEEQFKAEV 112
RF+ +L T+ +S ILG G FG V+KG L++G VAVK L G E QF+ EV
Sbjct: 118 RFSLRELQVATDSFSNKNILGRGGFGKVYKGRLADGSLVAVKRLKEERTPGGELQFQTEV 297
Query: 113 ITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRND-FDFQKLHKIAI 171
I H NL++L GFC +R LVY Y+ NGS+ + + D+ ++A+
Sbjct: 298 EMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPPYQEPLDWPTRKRVAL 477
Query: 172 GTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRG 231
G+A+G++YLH+ C +IIH D+K N+LLD + E + DFGLAKL + + + T RG
Sbjct: 478 GSARGLSYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYK-DTHVTTAVRG 654
Query: 232 TRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSS--YSESQQWFPRWTWEMFE 289
T G+ APE + K DV+ +GI+L E++ +R FD + ++ W + +
Sbjct: 655 TIGHIAPEYLSTGKSSEKTDVFGYGIMLLELITGQRAFDLARLANDDDVMLLDWVKGLLK 834
Query: 290 NNELVVMLALCEIEEKDSEI-AERMLKVALWCVQYSPNDRPLMSTVVKMLEGE 341
+L ML +++ E E++++VAL C Q SP DRP MS VV+MLEG+
Sbjct: 835 EKKL-EMLVDPDLQTNYIETEVEQLIQVALLCTQGSPMDRPKMSEVVRMLEGD 990
>TC217441 homologue to GB|AAK68074.1|14573459|AF384970 somatic embryogenesis
receptor-like kinase 3 {Arabidopsis thaliana;} , partial
(54%)
Length = 1008
Score = 173 bits (439), Expect = 1e-43
Identities = 107/292 (36%), Positives = 166/292 (56%), Gaps = 6/292 (2%)
Frame = +2
Query: 56 RFTPEKLDEITEKYST--ILGSGAFGVVFKGELSNGENVAVKVLNC-LDMGMEEQFKAEV 112
RF+ +L T+ +S ILG G FG V+KG L++G VAVK L G QF+ EV
Sbjct: 116 RFSLRELQVATDNFSNKHILGRGGFGKVYKGRLADGSLVAVKRLKEERTQGG*LQFQTEV 295
Query: 113 ITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFG-SKNRNDFDFQKLHKIAI 171
I H NL++L GFC +R LVY Y+ NGS+ + +++ + + +IA+
Sbjct: 296 EMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERQESQPPLGWPERKRIAL 475
Query: 172 GTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRG 231
G+A+G+AYLH+ C +IIH D+K N+LLD + E + DFGLAKL + + + T RG
Sbjct: 476 GSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYK-DTHVTTAVRG 652
Query: 232 TRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSS--YSESQQWFPRWTWEMFE 289
T G+ APE + K DV+ +G++L E++ +R FD + ++ W + +
Sbjct: 653 TIGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVKGLLK 832
Query: 290 NNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGE 341
+ +L ++ + E E++++VAL C Q SP +RP MS VV+MLEG+
Sbjct: 833 DRKLETLVDADLQGSYNDEEVEQLIQVALLCTQGSPMERPKMSEVVRMLEGD 988
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.134 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,055,993
Number of Sequences: 63676
Number of extensions: 230955
Number of successful extensions: 2956
Number of sequences better than 10.0: 975
Number of HSP's better than 10.0 without gapping: 2326
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2367
length of query: 402
length of database: 12,639,632
effective HSP length: 99
effective length of query: 303
effective length of database: 6,335,708
effective search space: 1919719524
effective search space used: 1919719524
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148970.5


