

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148969.6 + phase: 0
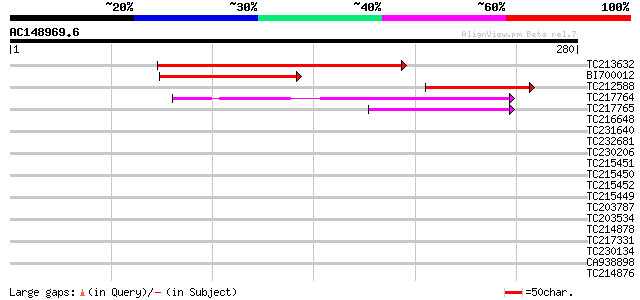
(280 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC213632 weakly similar to GB|AAG33973.1|11320952|AF250959 pepti... 135 2e-32
BI700012 similar to GP|11320952|gb| peptide deformylase-like pro... 98 5e-21
TC212588 similar to GB|AAG33973.1|11320952|AF250959 peptide defo... 84 5e-17
TC217764 similar to UP|DEFC_LYCES (Q9FV54) Peptide deformylase, ... 79 2e-15
TC217765 similar to GB|AAG33980.1|11320966|AF269165 peptide defo... 52 2e-07
TC216648 weakly similar to UP|Q9FFQ8 (Q9FFQ8) Emb|CAB62360.1, pa... 33 0.11
TC231640 homologue to UP|Q918P0 (Q918P0) Latency associated anti... 33 0.14
TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g4... 33 0.19
TC230206 similar to GB|AAP68317.1|31711922|BT008878 At5g64430 {A... 32 0.25
TC215451 weakly similar to UP|Q94K07 (Q94K07) Nucleosome assembl... 32 0.32
TC215450 similar to UP|P93488 (P93488) NAP1Ps, partial (30%) 32 0.32
TC215452 similar to UP|P93488 (P93488) NAP1Ps, partial (29%) 32 0.32
TC215449 similar to UP|P93488 (P93488) NAP1Ps, partial (97%) 32 0.32
TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%) 31 0.55
TC203534 31 0.71
TC214878 homologue to UP|RUBB_PEA (P08927) RuBisCO subunit bindi... 30 0.93
TC217331 similar to UP|Q9FH92 (Q9FH92) Emb|CAB62301.1 (At5g67210... 30 0.93
TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Pro... 30 0.93
CA938898 30 1.2
TC214876 homologue to UP|RUBB_PEA (P08927) RuBisCO subunit bindi... 30 1.2
>TC213632 weakly similar to GB|AAG33973.1|11320952|AF250959 peptide
deformylase-like protein {Arabidopsis thaliana;} ,
partial (38%)
Length = 464
Score = 135 bits (341), Expect = 2e-32
Identities = 71/123 (57%), Positives = 90/123 (72%)
Frame = +1
Query: 74 LKSELPHIVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGIP 133
L +L IV+AG+ VLH A EV+ EI S+++QKIID M+ VMR APG+ L+A +IGIP
Sbjct: 94 LAGKLAKIVKAGEAVLHSRAEEVEAIEIKSERVQKIIDDMVRVMRKAPGVGLAAPQIGIP 273
Query: 134 LRIIVLEEPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGFQ 193
LRIIVLE+ + + Y+ + K DR PFDLLVILNPKLK + +T LFFEGCLSV G+
Sbjct: 274 LRIIVLEDKIQYMAYYSNQELKAQDRTPFDLLVILNPKLKNTTTRTALFFEGCLSVPGYS 453
Query: 194 AVV 196
AVV
Sbjct: 454 AVV 462
>BI700012 similar to GP|11320952|gb| peptide deformylase-like protein
{Arabidopsis thaliana}, partial (34%)
Length = 420
Score = 97.8 bits (242), Expect = 5e-21
Identities = 47/70 (67%), Positives = 59/70 (84%)
Frame = +1
Query: 75 KSELPHIVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGIPL 134
K+ LP V+AGDPVLHEPA++VD +EI S+++QKIID MI VMR APG+ L+A +IGIPL
Sbjct: 190 KTNLPDTVKAGDPVLHEPAQDVDPNEIKSERVQKIIDDMIQVMRKAPGVGLAAPQIGIPL 369
Query: 135 RIIVLEEPKE 144
RIIVLE+ KE
Sbjct: 370 RIIVLEDTKE 399
>TC212588 similar to GB|AAG33973.1|11320952|AF250959 peptide deformylase-like
protein {Arabidopsis thaliana;} , partial (23%)
Length = 621
Score = 84.3 bits (207), Expect = 5e-17
Identities = 38/54 (70%), Positives = 46/54 (84%)
Frame = +3
Query: 206 GFDRYGEPIKINASGWHARILQHECDHLDGTLYVDKMVPRTFRSWENINMSIAR 259
G DRYG PIKI ASG ARIL HECDHLDGTLYVDKM+PRTFR+ +N+++ +A+
Sbjct: 3 GLDRYGAPIKIIASGCQARILPHECDHLDGTLYVDKMLPRTFRTVDNMDLPLAQ 164
>TC217764 similar to UP|DEFC_LYCES (Q9FV54) Peptide deformylase, chloroplast
precursor (PDF) (Polypeptide deformylase) , partial
(75%)
Length = 1880
Score = 79.3 bits (194), Expect = 2e-15
Identities = 53/169 (31%), Positives = 83/169 (48%)
Frame = +1
Query: 81 IVQAGDPVLHEPAREVDHSEINSDKIQKIIDGMILVMRNAPGISLSAQKIGIPLRIIVLE 140
IV+ DP L + + + D ++K++ M VM GI LSA ++GI ++++V
Sbjct: 277 IVEYPDPRLRARNKRIVAFD---DSLKKLVHEMFDVMYKTDGIGLSAPQLGINVQLMVF- 444
Query: 141 EPKENLYNYTEEVNKIIDRRPFDLLVILNPKLKIKSNKTFLFFEGCLSVHGFQAVVERYL 200
N + + + +V++NP++ S K LF EGCLS G A V+R
Sbjct: 445 -------------NPVGEHGEGEEIVLVNPRVSQYSKKLTLFNEGCLSFPGINADVKRPE 585
Query: 201 DVEVEGFDRYGEPIKINASGWHARILQHECDHLDGTLYVDKMVPRTFRS 249
V+++ D G +N S ARI QHE DHL G L+ ++M S
Sbjct: 586 SVKIDARDINGTRFSVNLSDLPARIFQHEFDHLQGILFFERMTEEVLDS 732
>TC217765 similar to GB|AAG33980.1|11320966|AF269165 peptide deformylase
{Arabidopsis thaliana;} , partial (41%)
Length = 682
Score = 52.4 bits (124), Expect = 2e-07
Identities = 29/72 (40%), Positives = 38/72 (52%)
Frame = +2
Query: 178 KTFLFFEGCLSVHGFQAVVERYLDVEVEGFDRYGEPIKINASGWHARILQHECDHLDGTL 237
K LF EGCLS G A V+R V+++ D G +N S ARI QHE D+L G L
Sbjct: 2 KLTLFNEGCLSFPGINADVKRPESVKIDARDINGTRFSVNLSDLPARIFQHEFDNLQGIL 181
Query: 238 YVDKMVPRTFRS 249
+ ++M S
Sbjct: 182 FFERMTEEVLDS 217
>TC216648 weakly similar to UP|Q9FFQ8 (Q9FFQ8) Emb|CAB62360.1, partial (41%)
Length = 2064
Score = 33.5 bits (75), Expect = 0.11
Identities = 18/38 (47%), Positives = 28/38 (73%)
Frame = -2
Query: 34 FSNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLS 71
FS+A+ SSS SSC++ + PS++ S FSSA+S ++ S
Sbjct: 389 FSSASSSSSFSSCSSFSSPSSSISSFFSSAASASSSTS 276
>TC231640 homologue to UP|Q918P0 (Q918P0) Latency associated antigen, partial
(7%)
Length = 466
Score = 33.1 bits (74), Expect = 0.14
Identities = 19/41 (46%), Positives = 27/41 (65%)
Frame = -3
Query: 36 NATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKS 76
N++VS SSS T T PS++ SSFSS+SS ++ S + S
Sbjct: 182 NSSVSCLSSSMITRTFPSSSSSSSFSSSSSSSSSSSSSSSS 60
Score = 29.3 bits (64), Expect = 2.1
Identities = 17/40 (42%), Positives = 25/40 (62%)
Frame = -3
Query: 26 RVVPMMLVFSNATVSSSSSSCNTETPPSNTKLSSFSSASS 65
R P S+ + SSSSSS ++ + S++ SSFSS+SS
Sbjct: 143 RTFPSSSSSSSFSSSSSSSSSSSSSSSSSSSSSSFSSSSS 24
Score = 28.1 bits (61), Expect = 4.6
Identities = 15/34 (44%), Positives = 24/34 (70%)
Frame = -3
Query: 40 SSSSSSCNTETPPSNTKLSSFSSASSETAFLSKT 73
SSSSSS ++ + S++ SS SS+SS ++F S +
Sbjct: 128 SSSSSSFSSSSSSSSSSSSSSSSSSSSSSFSSSS 27
>TC232681 weakly similar to GB|AAP21377.1|30102918|BT006569 At1g47970
{Arabidopsis thaliana;} , partial (39%)
Length = 867
Score = 32.7 bits (73), Expect = 0.19
Identities = 18/41 (43%), Positives = 27/41 (64%)
Frame = -2
Query: 38 TVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKSEL 78
++SSSSSS ++ +PPS SS SS+SS ++ SK + L
Sbjct: 311 SLSSSSSSSSSSSPPSGASSSSSSSSSSLSSSSSKDSRRRL 189
Score = 30.0 bits (66), Expect = 1.2
Identities = 16/34 (47%), Positives = 25/34 (73%)
Frame = -2
Query: 35 SNATVSSSSSSCNTETPPSNTKLSSFSSASSETA 68
S+++ SSSS S ++ +PP T LSS SS+SS ++
Sbjct: 374 SSSSSSSSSLSSSSGSPPRWTSLSSSSSSSSSSS 273
>TC230206 similar to GB|AAP68317.1|31711922|BT008878 At5g64430 {Arabidopsis
thaliana;} , partial (28%)
Length = 1305
Score = 32.3 bits (72), Expect = 0.25
Identities = 19/46 (41%), Positives = 27/46 (58%)
Frame = +1
Query: 34 FSNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKSELP 79
FS +T SSS C+ +PPS T ++ +S SS T+F +KT P
Sbjct: 259 FSPSTAPSSSPPCSPNSPPSATPQTTTTSPSS-TSFPAKTSTPSSP 393
>TC215451 weakly similar to UP|Q94K07 (Q94K07) Nucleosome assembly protein,
partial (20%)
Length = 777
Score = 32.0 bits (71), Expect = 0.32
Identities = 18/51 (35%), Positives = 34/51 (66%)
Frame = -1
Query: 30 MMLVFSNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKSELPH 80
++L+ S+++ SSSSSS ++ + S++ +SS SS+SS + S + S + H
Sbjct: 195 LVLLSSSSSSSSSSSSSSSSSSSSSSSISSSSSSSSRSPNSSP*MASPVNH 43
>TC215450 similar to UP|P93488 (P93488) NAP1Ps, partial (30%)
Length = 672
Score = 32.0 bits (71), Expect = 0.32
Identities = 18/51 (35%), Positives = 34/51 (66%)
Frame = -3
Query: 30 MMLVFSNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKSELPH 80
++L+ S+++ SSSSSS ++ + S++ +SS SS+SS + S + S + H
Sbjct: 316 LVLLSSSSSSSSSSSSSSSSSSSSSSSISSSSSSSSRSPNSSP*MASPVNH 164
Score = 27.3 bits (59), Expect = 7.9
Identities = 17/45 (37%), Positives = 27/45 (59%)
Frame = -3
Query: 32 LVFSNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKS 76
L F +SSSSSS ++ + S++ SS SS SS ++ S++ S
Sbjct: 328 LFFFLVLLSSSSSSSSSSSSSSSSSSSSSSSISSSSSSSSRSPNS 194
>TC215452 similar to UP|P93488 (P93488) NAP1Ps, partial (29%)
Length = 609
Score = 32.0 bits (71), Expect = 0.32
Identities = 18/51 (35%), Positives = 34/51 (66%)
Frame = -3
Query: 30 MMLVFSNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKSELPH 80
++L+ S+++ SSSSSS ++ + S++ +SS SS+SS + S + S + H
Sbjct: 244 LVLLSSSSSSSSSSSSSSSSSSSSSSSISSSSSSSSRSPNSSP*MASPVNH 92
>TC215449 similar to UP|P93488 (P93488) NAP1Ps, partial (97%)
Length = 1495
Score = 32.0 bits (71), Expect = 0.32
Identities = 18/51 (35%), Positives = 34/51 (66%)
Frame = -2
Query: 30 MMLVFSNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKSELPH 80
++L+ S+++ SSSSSS ++ + S++ +SS SS+SS + S + S + H
Sbjct: 1149 LVLLSSSSSSSSSSSSSSSSSSSSSSSISSSSSSSSRSPNSSP*MASPVNH 997
>TC203787 similar to UP|Q6VBJ3 (Q6VBJ3) Epa4p, partial (6%)
Length = 1056
Score = 31.2 bits (69), Expect = 0.55
Identities = 19/57 (33%), Positives = 30/57 (52%)
Frame = +3
Query: 36 NATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKSELPHIVQAGDPVLHEP 92
NA+ SSSSSS ++ + PS++ S SS+SS ++ S + P + P P
Sbjct: 300 NASSSSSSSSFSSSSAPSSSSSSPASSSSSSSSSSSSSSS*SSPSSSSSSSPFSSAP 470
>TC203534
Length = 1010
Score = 30.8 bits (68), Expect = 0.71
Identities = 26/96 (27%), Positives = 43/96 (44%), Gaps = 5/96 (5%)
Frame = +1
Query: 12 ETEMEAARRLASRLRVVPMMLVFSNATVSSSSSSCNTETPPSNTKLSSFSS-----ASSE 66
ETEM AA S R++P S+ ++ S +S TK SSF S A E
Sbjct: 358 ETEMIAAVGKGSSNRIIPDQAAASSDSIRSLGASA--------TKSSSFISPPIQGAEDE 513
Query: 67 TAFLSKTLKSELPHIVQAGDPVLHEPAREVDHSEIN 102
F+ K L+ + +++ + V H + ++ +N
Sbjct: 514 IKFIDKALEKGVVYMLAEAEVVAHPSSSILNKIVVN 621
>TC214878 homologue to UP|RUBB_PEA (P08927) RuBisCO subunit binding-protein
beta subunit, chloroplast precursor (60 kDa chaperonin
beta subunit) (CPN-60 beta), partial (26%)
Length = 687
Score = 30.4 bits (67), Expect = 0.93
Identities = 16/41 (39%), Positives = 23/41 (56%)
Frame = -3
Query: 25 LRVVPMMLVFSNATVSSSSSSCNTETPPSNTKLSSFSSASS 65
LR + FS ++VS + +C T TPP N + SFS + S
Sbjct: 493 LRASSTLNFFSLSSVSVCAPTCMTATPPDNLAILSFSFSFS 371
>TC217331 similar to UP|Q9FH92 (Q9FH92) Emb|CAB62301.1 (At5g67210), partial
(60%)
Length = 755
Score = 30.4 bits (67), Expect = 0.93
Identities = 23/79 (29%), Positives = 38/79 (47%), Gaps = 2/79 (2%)
Frame = +3
Query: 32 LVFSNATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKSELPHIVQAGDPVLHE 91
LVF + + ++ +T T S+T LSSF++ LS T+ + L H + H
Sbjct: 207 LVFLATLIYTRDTTSSTTTTTSSTPLSSFTN-----TLLSSTVINTLLHYASKSNDTFHM 371
Query: 92 PAREVDHSEIN--SDKIQK 108
P HS++ SD ++K
Sbjct: 372 P-----HSDLKTISDMLRK 413
>TC230134 weakly similar to UP|NUCL_HUMAN (P19338) Nucleolin (Protein C23),
partial (8%)
Length = 581
Score = 30.4 bits (67), Expect = 0.93
Identities = 15/29 (51%), Positives = 23/29 (78%)
Frame = -3
Query: 40 SSSSSSCNTETPPSNTKLSSFSSASSETA 68
SSSSSS ++ + PS++ LSS SS+SS ++
Sbjct: 516 SSSSSSSSSSSSPSSSSLSSSSSSSSSSS 430
Score = 30.0 bits (66), Expect = 1.2
Identities = 16/36 (44%), Positives = 27/36 (74%)
Frame = -3
Query: 36 NATVSSSSSSCNTETPPSNTKLSSFSSASSETAFLS 71
+++ SSSSSS ++ +P S++ SS SS+SS ++ LS
Sbjct: 525 SSSSSSSSSSSSSSSPSSSSLSSSSSSSSSSSSSLS 418
Score = 28.5 bits (62), Expect = 3.5
Identities = 16/36 (44%), Positives = 24/36 (66%)
Frame = -3
Query: 39 VSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTL 74
VSSSSSS ++ + S+ SS SS+SS ++ S +L
Sbjct: 528 VSSSSSSSSSSSSSSSPSSSSLSSSSSSSSSSSSSL 421
Score = 27.3 bits (59), Expect = 7.9
Identities = 16/38 (42%), Positives = 24/38 (63%)
Frame = -3
Query: 39 VSSSSSSCNTETPPSNTKLSSFSSASSETAFLSKTLKS 76
++S SSS + +PPS+ + SS SS+SS + S T S
Sbjct: 408 LASGSSSESASSPPSSPEKSSSSSSSSSPSSPSFTSSS 295
>CA938898
Length = 428
Score = 30.0 bits (66), Expect = 1.2
Identities = 21/90 (23%), Positives = 41/90 (45%), Gaps = 4/90 (4%)
Frame = +1
Query: 51 PPSNTKLSSFSSASSETAFLSKTLKSELPHIVQAGDPVLHEPAREVDHSEINSDKIQKII 110
PP+ + S ++ S + + T +P L AR VD ++ +Q ++
Sbjct: 118 PPAGSGFISGFTSISRRSSVDSTQNLSIPF---GPSSYLSAQARVVDEYSMSQIILQNVL 288
Query: 111 DG----MILVMRNAPGISLSAQKIGIPLRI 136
DG M++V+ A ++ ++ G+P RI
Sbjct: 289 DGGVTGMLIVVTGASHVTYGSRGTGVPARI 378
>TC214876 homologue to UP|RUBB_PEA (P08927) RuBisCO subunit binding-protein
beta subunit, chloroplast precursor (60 kDa chaperonin
beta subunit) (CPN-60 beta), partial (85%)
Length = 1797
Score = 30.0 bits (66), Expect = 1.2
Identities = 16/41 (39%), Positives = 23/41 (56%)
Frame = -1
Query: 25 LRVVPMMLVFSNATVSSSSSSCNTETPPSNTKLSSFSSASS 65
LR + FS ++VS + +C T TPP N + SFS + S
Sbjct: 1086 LRASSTLNFFSLSSVSVCAPTCITATPPDNLAILSFSFSFS 964
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.133 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,029,353
Number of Sequences: 63676
Number of extensions: 149621
Number of successful extensions: 1784
Number of sequences better than 10.0: 86
Number of HSP's better than 10.0 without gapping: 1451
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1616
length of query: 280
length of database: 12,639,632
effective HSP length: 96
effective length of query: 184
effective length of database: 6,526,736
effective search space: 1200919424
effective search space used: 1200919424
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148969.6


