

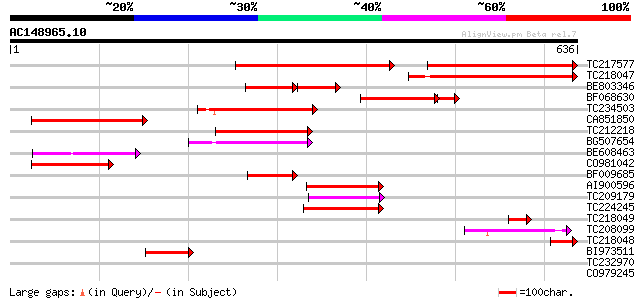
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148965.10 + phase: 0
(636 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC217577 similar to UP|Q9ASS7 (Q9ASS7) AT5g36940/MLF18_60, parti... 262 e-115
TC218047 similar to UP|Q8GT81 (Q8GT81) Fiber protein Fb12 (Fragm... 306 1e-83
BE803346 117 3e-46
BF068630 120 7e-29
TC234503 similar to PIR|T48473|T48473 amino acid transport-like ... 108 9e-24
CA851850 weakly similar to PIR|T48473|T48 amino acid transport-l... 106 4e-23
TC212218 similar to PIR|T48473|T48473 amino acid transport-like ... 102 5e-22
BG507654 96 4e-20
BE608463 similar to PIR|T48473|T48 amino acid transport-like pro... 85 1e-16
CO981042 81 2e-15
BF009685 79 6e-15
AI900596 weakly similar to GP|13605811|gb| AT5g36940/MLF18_60 {A... 77 2e-14
TC209179 similar to PIR|T48473|T48473 amino acid transport-like ... 63 5e-10
TC224245 similar to PIR|A86307|A86307 amino acid transporter hom... 60 4e-09
TC218049 similar to GB|AAP37728.1|30725412|BT008369 At3g03720 {A... 59 7e-09
TC208099 similar to UP|Q6H6H8 (Q6H6H8) Amino acid permease-like,... 57 3e-08
TC218048 similar to UP|Q8GT81 (Q8GT81) Fiber protein Fb12 (Fragm... 54 3e-07
BI973511 42 8e-04
TC232970 similar to PIR|T48473|T48473 amino acid transport-like ... 38 0.012
CO979245 29 5.6
>TC217577 similar to UP|Q9ASS7 (Q9ASS7) AT5g36940/MLF18_60, partial (44%)
Length = 1475
Score = 262 bits (669), Expect(2) = e-115
Identities = 128/178 (71%), Positives = 154/178 (85%)
Frame = +3
Query: 254 IGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPYYELNPDTPISSA 313
IGFD+V STAEEVKNPQRDLP+GI +L +CC LYMLVS VIVGLVPYY +NPDTPISSA
Sbjct: 6 IGFDAVASTAEEVKNPQRDLPLGIGGSLFLCCGLYMLVSIVIVGLVPYYAINPDTPISSA 185
Query: 314 FSSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMARDGLLPTFFSDIHRRTQIPL 373
F+ GM+WA Y+I GA TAL +SL+G +LPQPR+ MAMARDGLLP FFSDI++ +Q+P+
Sbjct: 186 FADNGMQWAAYVINGGAFTALCASLMGGILPQPRILMAMARDGLLPPFFSDINKCSQVPV 365
Query: 374 KSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVLIIRYVPPDEIPIPASL 431
KSTIVTGL A++LAF M+VS+LAGMVSVGTLLAFT VA+SVLI+RY+PPDE+ + SL
Sbjct: 366 KSTIVTGLVASLLAFSMEVSELAGMVSVGTLLAFTMVAISVLILRYIPPDEVLLLPSL 539
Score = 172 bits (436), Expect(2) = e-115
Identities = 81/169 (47%), Positives = 114/169 (66%), Gaps = 1/169 (0%)
Frame = +1
Query: 469 KSDVLLEHPLIIKE-VTKEQHNEKTRRKLAAWTIALLCIGILIVSGSASAERCPRILRVT 527
K DV ++ PLI K+ V + +E RRK+ W IA C+G+ +++ +AS + +R
Sbjct: 655 KEDVPIDCPLIAKDPVIGKYVHEGNRRKVIGWVIAFTCLGVFVLTFAASNKTMISSVRFA 834
Query: 528 LFGAGVVIFLCSIIVLACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVD 587
L G G + L + L C+ QDD RH FGHSGGF CPFVP LP ACILIN+YLL++LG
Sbjct: 835 LCGVGGCLLLSGFVFLTCMDQDDARHNFGHSGGFTCPFVPLLPVACILINSYLLVNLGAA 1014
Query: 588 TWLRVSVWLLIGVLIYLFYGRTHSSLLNAIYVPSARADEIHRSQANHLA 636
TW RVS+WL IGV++Y+FYGRTHS+L +A+ VP+ + +I+ + + LA
Sbjct: 1015TWARVSIWLAIGVIVYVFYGRTHSTLKDAVCVPATQVVDIYHTSTSCLA 1161
>TC218047 similar to UP|Q8GT81 (Q8GT81) Fiber protein Fb12 (Fragment),
partial (91%)
Length = 833
Score = 306 bits (785), Expect = 1e-83
Identities = 151/189 (79%), Positives = 169/189 (88%)
Frame = +1
Query: 448 EDRTVSPVDLASYSDNSHLHDKSDVLLEHPLIIKEVTKEQHNEKTRRKLAAWTIALLCIG 507
ED +VSPVD ASY +NSHL + LL HPLIIKEVTK++ NEK RRKLA+WTIALLCIG
Sbjct: 7 EDGSVSPVDPASYCENSHL----EALLGHPLIIKEVTKDEQNEKARRKLASWTIALLCIG 174
Query: 508 ILIVSGSASAERCPRILRVTLFGAGVVIFLCSIIVLACIKQDDTRHTFGHSGGFACPFVP 567
ILIV+ +ASA+ CPRILR+TL G G ++ LCS IVLAC+KQDDTRH+FGHSGGFACPFVP
Sbjct: 175 ILIVTSAASADWCPRILRLTLCGMGGILLLCSTIVLACVKQDDTRHSFGHSGGFACPFVP 354
Query: 568 FLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLIYLFYGRTHSSLLNAIYVPSARADEI 627
FLPAACILINTYLLIDLGV TWLRVSVW+LIGVL+YLFYGRTHSSLL+AIYVPSA ADEI
Sbjct: 355 FLPAACILINTYLLIDLGVATWLRVSVWMLIGVLVYLFYGRTHSSLLHAIYVPSAYADEI 534
Query: 628 HRSQANHLA 636
HRSQA+HLA
Sbjct: 535 HRSQASHLA 561
>BE803346
Length = 437
Score = 117 bits (293), Expect(2) = 3e-46
Identities = 55/59 (93%), Positives = 58/59 (98%)
Frame = +3
Query: 265 EVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPYYELNPDTPISSAFSSYGMEWAV 323
+VKNPQRDLPIGISTAL ICCVLYMLV+AVIVGLVPYYELNPDTPISSAFSSYGM+WAV
Sbjct: 18 QVKNPQRDLPIGISTALTICCVLYMLVAAVIVGLVPYYELNPDTPISSAFSSYGMQWAV 194
Score = 87.0 bits (214), Expect(2) = 3e-46
Identities = 43/48 (89%), Positives = 46/48 (95%)
Frame = +1
Query: 324 YIITTGAVTALFSSLLGSVLPQPRVFMAMARDGLLPTFFSDIHRRTQI 371
YIITTGAVTALF+SLLGSVLPQPRVFMAMARDGLLP FFS+IH+ TQI
Sbjct: 292 YIITTGAVTALFASLLGSVLPQPRVFMAMARDGLLPHFFSNIHKGTQI 435
>BF068630
Length = 336
Score = 120 bits (301), Expect(2) = 7e-29
Identities = 62/89 (69%), Positives = 72/89 (80%)
Frame = +2
Query: 394 QLAGMVSVGTLLAFTTVAVSVLIIRYVPPDEIPIPASLLTSVDPLLRHSGDDIEEDRTVS 453
QLAGMVSVGTLLAFTTVAVSVLIIRYVPPDE+P+ +SLLTSVDPLLRHSG DI ED +
Sbjct: 2 QLAGMVSVGTLLAFTTVAVSVLIIRYVPPDEVPVLSSLLTSVDPLLRHSGGDIGEDGAII 181
Query: 454 PVDLASYSDNSHLHDKSDVLLEHPLIIKE 482
VD SY +N ++H K + ++ PLI KE
Sbjct: 182 YVDSFSYCENRYVHHK*EPVIGDPLIFKE 268
Score = 25.8 bits (55), Expect(2) = 7e-29
Identities = 12/25 (48%), Positives = 16/25 (64%)
Frame = +3
Query: 480 IKEVTKEQHNEKTRRKLAAWTIALL 504
+K ++ + N RKLA WTIALL
Sbjct: 261 LKNLS*DDDNGIAGRKLATWTIALL 335
>TC234503 similar to PIR|T48473|T48473 amino acid transport-like protein -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(24%)
Length = 424
Score = 108 bits (269), Expect = 9e-24
Identities = 56/143 (39%), Positives = 92/143 (64%), Gaps = 8/143 (5%)
Frame = +1
Query: 211 FIIIVGGYLGFKAGWVGY------ELPSGYFPYGVNGMFAGSAIVFFSYIGFDSVTSTAE 264
F+I++G + G W + PSG+FP+G G+F G+A+V+ SYIG+D+V++ AE
Sbjct: 1 FVIMMGFWRG---NWKNFTEPANPHNPSGFFPHGAAGVFKGAALVYLSYIGYDAVSTMAE 171
Query: 265 EVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPYYELNPDTPISSAFS--SYGMEWA 322
EV++P +D+P+G+S ++ + VLY L++A + L+PY +N + P S+AFS S G W
Sbjct: 172 EVRDPVKDIPVGVSGSVVVVTVLYCLMAASMTKLLPYDVINAEAPFSAAFSGRSDGWGWV 351
Query: 323 VYIITTGAVTALFSSLLGSVLPQ 345
+I GA + +SLL ++L Q
Sbjct: 352 SRVIGVGASFGILTSLLVAMLGQ 420
>CA851850 weakly similar to PIR|T48473|T48 amino acid transport-like protein
- Arabidopsis thaliana, partial (20%)
Length = 617
Score = 106 bits (264), Expect = 4e-23
Identities = 51/130 (39%), Positives = 79/130 (60%)
Frame = +3
Query: 25 KQVDSIHYRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISLFIAG 84
++ + + +R + RKL DLV +GVG +G GV++ G+VA +GP++ IS IAG
Sbjct: 201 QEFNQVRHRSGADMKRKLKWHDLVALGVGGMLGVGVFVTTGSVALHHSGPSVFISYIIAG 380
Query: 85 IAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVARGITP 144
I+A LS+LCY E A + P AG A+ Y + GE + + G ++++EY +AVAR T
Sbjct: 381 ISALLSSLCYTEFAVQVPVAGGAFSYLRLTFGEFLGYFAGANILMEYVFSNAAVARSFTE 560
Query: 145 NLALFFGGQD 154
L++ FG D
Sbjct: 561 YLSIAFGEND 590
>TC212218 similar to PIR|T48473|T48473 amino acid transport-like protein -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(22%)
Length = 401
Score = 102 bits (254), Expect = 5e-22
Identities = 49/111 (44%), Positives = 77/111 (69%), Gaps = 2/111 (1%)
Frame = +1
Query: 231 PSGYFPYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYML 290
P+G+FP+G G+F G+A V+ SYIG+D+V++ AEEV++P +D+P+G+S ++ I VLY L
Sbjct: 64 PTGFFPHGAAGVFKGAAAVYLSYIGYDAVSTMAEEVRDPVKDIPVGVSGSVVIVTVLYCL 243
Query: 291 VSAVIVGLVPYYELNPDTPISSAFS--SYGMEWAVYIITTGAVTALFSSLL 339
++A + L+PY +N + P S+AFS S G W +I GA + +SLL
Sbjct: 244 MAASMTKLLPYDMINAEAPFSAAFSGRSDGWGWISGVIGVGASFGILTSLL 396
>BG507654
Length = 411
Score = 96.3 bits (238), Expect = 4e-20
Identities = 54/139 (38%), Positives = 79/139 (55%)
Frame = +3
Query: 201 VTTINVSVMLFIIIVGGYLGFKAGWVGYELPSGYFPYGVNGMFAGSAIVFFSYIGFDSVT 260
+T +V FIII G G V P G P+G G+ G+AIV+FSYIG+DS +
Sbjct: 3 MTAFHVIFFGFIIIAGYCNGSAKNLVS---PKGLAPFGARGVLDGAAIVYFSYIGYDSAS 173
Query: 261 STAEEVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPYYELNPDTPISSAFSSYGME 320
+ AEEVK+P + LPIGI ++ I +LY L++ + +VPY +++ S AF G
Sbjct: 174 TMAEEVKDPFKSLPIGIVGSVLITTLLYCLMALSLCMMVPYNKISEKASFSIAFLKIGWN 353
Query: 321 WAVYIITTGAVTALFSSLL 339
WA ++ GA + +SLL
Sbjct: 354 WASNLVGAGASLGIVASLL 410
>BE608463 similar to PIR|T48473|T48 amino acid transport-like protein -
Arabidopsis thaliana, partial (25%)
Length = 463
Score = 84.7 bits (208), Expect = 1e-16
Identities = 46/121 (38%), Positives = 67/121 (55%)
Frame = +2
Query: 26 QVDSIHYRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISLFIAGI 85
++ + R + + L DLVG+ +G +GAGV++ G R AGPA+++S IAG
Sbjct: 92 EMSQVRSRSGTSMRKTLRWFDLVGLSIGGMVGAGVFVTTGHATR-YAGPAVLLSYAIAGF 268
Query: 86 AAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVARGITPN 145
A LS CY E A P AG A+ Y I GE A+L G +L+ +Y + +AVA T +
Sbjct: 269 CALLSGFCYTEFAVDMPVAGGAFSYLRITCGEYAAFLTGANLVEDYVLSNAAVAISWTGD 448
Query: 146 L 146
L
Sbjct: 449 L 451
>CO981042
Length = 666
Score = 80.9 bits (198), Expect = 2e-15
Identities = 38/92 (41%), Positives = 57/92 (61%)
Frame = +2
Query: 25 KQVDSIHYRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISLFIAG 84
++ + + +R + RKL DLV +GVG +G GV++ G+VA +GP++ IS IAG
Sbjct: 176 QEFNQVRHRSGADMKRKLKWHDLVALGVGGMLGVGVFVTTGSVALHHSGPSVFISYIIAG 355
Query: 85 IAAALSALCYAELACRCPSAGSAYHYTYICIG 116
I+A LS+LCY E A + P AG A+ Y + G
Sbjct: 356 ISALLSSLCYTEFAVQVPVAGGAFSYLRLTFG 451
>BF009685
Length = 169
Score = 79.0 bits (193), Expect = 6e-15
Identities = 37/56 (66%), Positives = 44/56 (78%)
Frame = +2
Query: 267 KNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPYYELNPDTPISSAFSSYGMEWA 322
+NP RDLP+GI A ICC+L MLVSA +VGL PY +LN DTPI SAFS+Y M+WA
Sbjct: 2 RNPDRDLPVGI*IA*TICCILSMLVSADLVGLHPYDKLNSDTPICSAFSTYAMQWA 169
>AI900596 weakly similar to GP|13605811|gb| AT5g36940/MLF18_60 {Arabidopsis
thaliana}, partial (10%)
Length = 259
Score = 77.4 bits (189), Expect = 2e-14
Identities = 44/86 (51%), Positives = 60/86 (69%)
Frame = +2
Query: 334 LFSSLLGSVLPQPRVFMAMARDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVS 393
L +SL+G +L + MAMA DGLL F DI+ +P+ ST+V+GL ++LAF M VS
Sbjct: 2 LCASLMGVILSHS*ILMAMAMDGLLSPLFFDIY*SILLPV*STLVSGLVFSLLAFSMVVS 181
Query: 394 QLAGMVSVGTLLAFTTVAVSVLIIRY 419
+L+ MVSV TLLAFT VA+ +LI+ Y
Sbjct: 182 ELSSMVSVCTLLAFTMVALFLLILTY 259
>TC209179 similar to PIR|T48473|T48473 amino acid transport-like protein -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(36%)
Length = 921
Score = 62.8 bits (151), Expect = 5e-10
Identities = 30/85 (35%), Positives = 51/85 (59%)
Frame = +1
Query: 336 SSLLGSVLPQPRVFMAMARDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQL 395
+SLL ++L Q R + R ++P++F+ +H +T P+ ++ G+F A +A F D+ L
Sbjct: 1 ASLLVAMLGQARYMCVIGRSNVVPSWFARVHPKTSTPVNASAFLGIFTAAIALFTDLDVL 180
Query: 396 AGMVSVGTLLAFTTVAVSVLIIRYV 420
+V +GTL F VA +V+ RYV
Sbjct: 181 FNLVCIGTLFVFYMVANAVIYRRYV 255
Score = 37.7 bits (86), Expect = 0.016
Identities = 17/49 (34%), Positives = 30/49 (60%)
Frame = +1
Query: 564 PFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLIYLFYGRTHSS 612
PF+P++P+ I +N +LL L +++R + + VL Y+FY H+S
Sbjct: 466 PFMPWIPSISIFLNVFLLGSLDGPSYVRFGFFSAVAVLFYVFYS-VHAS 609
>TC224245 similar to PIR|A86307|A86307 amino acid transporter homolog
[imported] - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (34%)
Length = 843
Score = 59.7 bits (143), Expect = 4e-09
Identities = 29/90 (32%), Positives = 55/90 (60%)
Frame = +3
Query: 330 AVTALFSSLLGSVLPQPRVFMAMARDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFF 389
A+ + +SLL + Q R +AR ++P FF+ +H +T P+ +T++T + ++V+A F
Sbjct: 3 ALKGMTTSLLVGSMGQARYTTQIARSHMIPPFFALVHPKTGTPVNATLLTTISSSVIALF 182
Query: 390 MDVSQLAGMVSVGTLLAFTTVAVSVLIIRY 419
+ L+ + S+ TL F +AV++L+ RY
Sbjct: 183 SSLDVLSSVFSISTLFIFMLMAVALLVRRY 272
>TC218049 similar to GB|AAP37728.1|30725412|BT008369 At3g03720 {Arabidopsis
thaliana;} , partial (4%)
Length = 602
Score = 58.9 bits (141), Expect = 7e-09
Identities = 26/26 (100%), Positives = 26/26 (100%)
Frame = +2
Query: 560 GFACPFVPFLPAACILINTYLLIDLG 585
GFACPFVPFLPAACILINTYLLIDLG
Sbjct: 479 GFACPFVPFLPAACILINTYLLIDLG 556
>TC208099 similar to UP|Q6H6H8 (Q6H6H8) Amino acid permease-like, partial
(16%)
Length = 769
Score = 56.6 bits (135), Expect = 3e-08
Identities = 40/131 (30%), Positives = 62/131 (46%), Gaps = 11/131 (8%)
Frame = +1
Query: 511 VSGSASAERCPRILRVTL--FGAGVV-------IFLCSIIVLACIKQDDT--RHTFGHSG 559
VS SA E ++ V L F +G++ IFL +V+A R + +
Sbjct: 55 VSSSAEREGIICLIAVALCGFASGLLYRYDASFIFLILALVIAAGASAALVFRQVYADAP 234
Query: 560 GFACPFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLIYLFYGRTHSSLLNAIYV 619
GF+CP VP LP CI N +L L + W+R + ++ V +Y YG+ H++
Sbjct: 235 GFSCPGVPLLPNICIFFNMFLFAQLHHEAWVRFVILCVVMVGVYAIYGQYHAN------- 393
Query: 620 PSARADEIHRS 630
PSA + HR+
Sbjct: 394 PSAEENVYHRA 426
>TC218048 similar to UP|Q8GT81 (Q8GT81) Fiber protein Fb12 (Fragment),
partial (21%)
Length = 323
Score = 53.5 bits (127), Expect = 3e-07
Identities = 26/30 (86%), Positives = 28/30 (92%)
Frame = +2
Query: 607 GRTHSSLLNAIYVPSARADEIHRSQANHLA 636
GR+HSSLL+AIYVPSA ADEIHRSQA HLA
Sbjct: 2 GRSHSSLLHAIYVPSAYADEIHRSQAIHLA 91
>BI973511
Length = 421
Score = 42.0 bits (97), Expect = 8e-04
Identities = 19/54 (35%), Positives = 34/54 (62%)
Frame = +1
Query: 153 QDNLPSFLARHTLPGLGIVVDPCAAVLIVLITLLLCLGIKESSTVQSIVTTINV 206
+DN+P ++ G + ++ A +L+VL+T +LC G++ESS V S++T V
Sbjct: 7 KDNIPKWIGHGEDIGDVLSINVLAPILLVLLTFILCRGVQESSVVNSLMTVTKV 168
Score = 40.0 bits (92), Expect = 0.003
Identities = 20/74 (27%), Positives = 38/74 (51%), Gaps = 2/74 (2%)
Frame = +2
Query: 188 CLGIKES--STVQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYELPSGYFPYGVNGMFAG 245
C G+ E S + + + +++ +I G + + W S + P G+ +F G
Sbjct: 218 C*GLSERLRSNDKLFYCLLQIIIVIIVIFAGAFEVDVSNW------SPFAPNGLKAIFTG 379
Query: 246 SAIVFFSYIGFDSV 259
+ +VFF+Y+GFD+V
Sbjct: 380 ATVVFFAYVGFDAV 421
>TC232970 similar to PIR|T48473|T48473 amino acid transport-like protein -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(9%)
Length = 888
Score = 38.1 bits (87), Expect = 0.012
Identities = 14/47 (29%), Positives = 29/47 (60%)
Frame = +3
Query: 561 FACPFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLIYLFYG 607
++ PF+P+ PA I +N +L+ L + ++ R ++W + + Y+ YG
Sbjct: 309 WSVPFMPWPPAMSIFLNVFLMTTLKILSFQRFAIWACLITIFYVLYG 449
Score = 30.4 bits (67), Expect = 2.5
Identities = 14/43 (32%), Positives = 24/43 (55%)
Frame = +3
Query: 384 AVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVLIIRYVPPDEIP 426
A + F ++ + ++S+GTLL F VA +++ RYV P
Sbjct: 3 ATITLFTELDIIIELISIGTLLVFYMVANALIYRRYVITSHAP 131
>CO979245
Length = 730
Score = 29.3 bits (64), Expect = 5.6
Identities = 18/43 (41%), Positives = 22/43 (50%)
Frame = -1
Query: 220 GFKAGWVGYELPSGYFPYGVNGMFAGSAIVFFSYIGFDSVTST 262
GF G+ G PSG+ + G FAG A + GF SV ST
Sbjct: 487 GFGGGFAGSSSPSGFSNTAIGGGFAGIA---STGRGFGSVAST 368
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.142 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,473,313
Number of Sequences: 63676
Number of extensions: 457407
Number of successful extensions: 3170
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 3111
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3165
length of query: 636
length of database: 12,639,632
effective HSP length: 103
effective length of query: 533
effective length of database: 6,081,004
effective search space: 3241175132
effective search space used: 3241175132
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148965.10


