

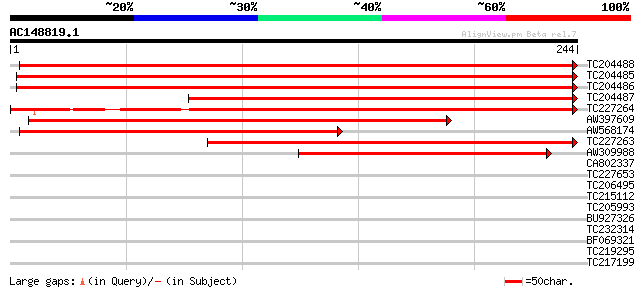
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148819.1 + phase: 0
(244 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204488 similar to PRF|1909359B.0|445613|1909359B ribosomal pro... 436 e-123
TC204485 similar to UP|RL73_ARATH (Q9LHP1) 60S ribosomal protein... 433 e-122
TC204486 similar to UP|RL73_ARATH (Q9LHP1) 60S ribosomal protein... 430 e-121
TC204487 similar to PRF|1909359B.0|445613|1909359B ribosomal pro... 322 1e-88
TC227264 weakly similar to PRF|1909359B.0|445613|1909359B riboso... 221 2e-58
AW397609 similar to GP|11994560|db 60S ribosomal protein L7 {Ara... 221 3e-58
AW568174 similar to GP|11994560|db 60S ribosomal protein L7 {Ara... 197 5e-51
TC227263 weakly similar to UP|RL7_DICDI (P11874) 60S ribosomal p... 186 6e-48
AW309988 weakly similar to GP|6687301|emb| 60S ribosomal protein... 101 3e-22
CA802337 32 0.26
TC227653 similar to UP|O80383 (O80383) 98b, partial (78%) 31 0.45
TC206495 weakly similar to GB|AAO42927.1|28416793|BT004681 At5g2... 28 2.9
TC215112 28 2.9
TC205993 weakly similar to UP|Q6ZC61 (Q6ZC61) Ribosomal protein ... 28 2.9
BU927326 28 3.8
TC232314 weakly similar to UP|Q6NLY8 (Q6NLY8) At4g36720, partial... 28 3.8
BF069321 28 3.8
TC219295 28 4.9
TC217199 weakly similar to UP|Q7X9D9 (Q7X9D9) ABRH1 (Fragment), ... 27 8.4
>TC204488 similar to PRF|1909359B.0|445613|1909359B ribosomal protein L7.
{Solanum tuberosum;} , complete
Length = 936
Score = 436 bits (1121), Expect = e-123
Identities = 212/240 (88%), Positives = 230/240 (95%)
Frame = +2
Query: 5 EVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQKEL 64
E K +VPESV+KK+KRNEEWAL KKQE ++AKK R ETRKLI++RAKQY+KEYD+ QKEL
Sbjct: 38 EAKALVPESVVKKEKRNEEWALAKKQELDAAKKMRFETRKLIFNRAKQYSKEYDEQQKEL 217
Query: 65 IALKREAKLKGGFYVDPEAKLLFIIRIRGINAMDPKSRKILQLLRLRQICNGVFLKVNKA 124
I LKREAKLKGGFYV+PEAKLLFIIRIRGINAMDPK+RKILQLLRLRQI NGVFLKVNKA
Sbjct: 218 IRLKREAKLKGGFYVEPEAKLLFIIRIRGINAMDPKTRKILQLLRLRQIFNGVFLKVNKA 397
Query: 125 TVNMLHRVEPYVTYGYPNLKSVRELIYKRGYGKVDRQRIALTDNSIIEQVLGKHGIICIE 184
TVNMLHRVEPYVTYGYPNLKSVRELIYKRGYGKV++QRIALTDNSIIEQ LGKHGIIC+E
Sbjct: 398 TVNMLHRVEPYVTYGYPNLKSVRELIYKRGYGKVNKQRIALTDNSIIEQTLGKHGIICVE 577
Query: 185 DLIHEILTVGPHFREANNFLWPFKLKAPLGGMKKKRNHYVEGGDAGNREDYINELIRRMN 244
DLIHEI+TVGPHF+EANNFLWPFKLKAPLGG++KKRNHYVEGGDAGNREDYINELIRRMN
Sbjct: 578 DLIHEIMTVGPHFKEANNFLWPFKLKAPLGGLEKKRNHYVEGGDAGNREDYINELIRRMN 757
>TC204485 similar to UP|RL73_ARATH (Q9LHP1) 60S ribosomal protein L7-3,
partial (98%)
Length = 1025
Score = 433 bits (1113), Expect = e-122
Identities = 213/241 (88%), Positives = 229/241 (94%)
Frame = +1
Query: 4 EEVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQKE 63
EEVK VVPESVLKKQKR EEWAL KKQ+ E+AKKKR+E+RKLI++RAKQYAKEYD +KE
Sbjct: 76 EEVKAVVPESVLKKQKREEEWALAKKQDLEAAKKKRAESRKLIYNRAKQYAKEYDHQEKE 255
Query: 64 LIALKREAKLKGGFYVDPEAKLLFIIRIRGINAMDPKSRKILQLLRLRQICNGVFLKVNK 123
LI LKREAKLKGGFYVDPEAKLLFI+RIRGINAMDPKSRKILQLLRLRQI NGVFLKVNK
Sbjct: 256 LIRLKREAKLKGGFYVDPEAKLLFIVRIRGINAMDPKSRKILQLLRLRQIFNGVFLKVNK 435
Query: 124 ATVNMLHRVEPYVTYGYPNLKSVRELIYKRGYGKVDRQRIALTDNSIIEQVLGKHGIICI 183
ATVNMLHRVEPYVTYGYPNLKSVRELIYKRGYGK+++QR ALTDNSIIEQ LGK+GII
Sbjct: 436 ATVNMLHRVEPYVTYGYPNLKSVRELIYKRGYGKLNKQRTALTDNSIIEQALGKYGIIST 615
Query: 184 EDLIHEILTVGPHFREANNFLWPFKLKAPLGGMKKKRNHYVEGGDAGNREDYINELIRRM 243
EDLIHEI+TVGPHF+EANNFLWPFKLKAPLGG+KKKRNHYVEGGDAGNRE+YINELIRRM
Sbjct: 616 EDLIHEIITVGPHFKEANNFLWPFKLKAPLGGLKKKRNHYVEGGDAGNRENYINELIRRM 795
Query: 244 N 244
N
Sbjct: 796 N 798
>TC204486 similar to UP|RL73_ARATH (Q9LHP1) 60S ribosomal protein L7-3,
partial (98%)
Length = 1021
Score = 430 bits (1106), Expect = e-121
Identities = 213/241 (88%), Positives = 228/241 (94%)
Frame = +3
Query: 4 EEVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQKE 63
EEVK VVPESVLKK+KR EEWAL KKQE ++AKKKR+E+RKLI++RAKQYAKEYD +KE
Sbjct: 69 EEVKVVVPESVLKKRKREEEWALAKKQELDAAKKKRAESRKLIYNRAKQYAKEYDHQEKE 248
Query: 64 LIALKREAKLKGGFYVDPEAKLLFIIRIRGINAMDPKSRKILQLLRLRQICNGVFLKVNK 123
LI LKREAKLKGGFYVDPEAKLLFIIRIRGINAMDPKSRKILQLLRLRQI NGVFLKVNK
Sbjct: 249 LIRLKREAKLKGGFYVDPEAKLLFIIRIRGINAMDPKSRKILQLLRLRQIFNGVFLKVNK 428
Query: 124 ATVNMLHRVEPYVTYGYPNLKSVRELIYKRGYGKVDRQRIALTDNSIIEQVLGKHGIICI 183
ATVNMLHRVEPYVTYGYPNLKSVRELIYKRGYGK+ +QR ALTDNSIIEQ LGK+GII
Sbjct: 429 ATVNMLHRVEPYVTYGYPNLKSVRELIYKRGYGKLMKQRTALTDNSIIEQALGKYGIIST 608
Query: 184 EDLIHEILTVGPHFREANNFLWPFKLKAPLGGMKKKRNHYVEGGDAGNREDYINELIRRM 243
EDLIHEI+TVGPHF+EANNFLWPFKLKAPLGG+KKKRNHYVEGGDAGNRE+YINELIRRM
Sbjct: 609 EDLIHEIITVGPHFKEANNFLWPFKLKAPLGGLKKKRNHYVEGGDAGNRENYINELIRRM 788
Query: 244 N 244
N
Sbjct: 789 N 791
>TC204487 similar to PRF|1909359B.0|445613|1909359B ribosomal protein L7.
{Solanum tuberosum;} , partial (69%)
Length = 685
Score = 322 bits (825), Expect = 1e-88
Identities = 154/167 (92%), Positives = 164/167 (97%)
Frame = +3
Query: 78 YVDPEAKLLFIIRIRGINAMDPKSRKILQLLRLRQICNGVFLKVNKATVNMLHRVEPYVT 137
YV+PEAKLLFIIRIRGINAMDPK+RKILQLLRLRQI NGVFLKVNKATVNMLHRVEPYVT
Sbjct: 3 YVEPEAKLLFIIRIRGINAMDPKTRKILQLLRLRQIFNGVFLKVNKATVNMLHRVEPYVT 182
Query: 138 YGYPNLKSVRELIYKRGYGKVDRQRIALTDNSIIEQVLGKHGIICIEDLIHEILTVGPHF 197
YGYPNLKSV+ELIYKRGYGKV++QRIAL DNSI+EQ LGKHGIIC+EDLIHEILTVGPHF
Sbjct: 183 YGYPNLKSVKELIYKRGYGKVNKQRIALNDNSIVEQTLGKHGIICVEDLIHEILTVGPHF 362
Query: 198 REANNFLWPFKLKAPLGGMKKKRNHYVEGGDAGNREDYINELIRRMN 244
+EANNFLWPFKLKAPLGG++KKRNHYVEGGDAGNREDYINELIRRMN
Sbjct: 363 KEANNFLWPFKLKAPLGGLEKKRNHYVEGGDAGNREDYINELIRRMN 503
>TC227264 weakly similar to PRF|1909359B.0|445613|1909359B ribosomal protein
L7. {Solanum tuberosum;} , partial (44%)
Length = 1105
Score = 221 bits (564), Expect = 2e-58
Identities = 113/246 (45%), Positives = 165/246 (66%), Gaps = 2/246 (0%)
Frame = +3
Query: 1 MAKEEVKTV--VPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYD 58
MA+E+ K + + E +LKK+K E WAL +K+EQ KK +S + + + EY
Sbjct: 63 MAEEDSKPLNYISEVILKKRKNTEAWAL-RKKEQFQQKKYQSI------KKPEDFIHEYR 221
Query: 59 DSQKELIALKREAKLKGGFYVDPEAKLLFIIRIRGINAMDPKSRKILQLLRLRQICNGVF 118
+ + +LI +KR K K + + + IIRI+G M P++RK+L L LR+ + VF
Sbjct: 222 NKEVDLIRMKRRVKRK---VPEANSNTIIIIRIQGKKDMHPRTRKVLYSLGLRRRFSAVF 392
Query: 119 LKVNKATVNMLHRVEPYVTYGYPNLKSVRELIYKRGYGKVDRQRIALTDNSIIEQVLGKH 178
+K ++ + L RVEPYVTYGYPNLKS++ELIYK+G+ +++++++ LTDN+IIEQ LGK
Sbjct: 393 VKPSEGIMAKLQRVEPYVTYGYPNLKSIKELIYKKGHARMEQRKVPLTDNNIIEQELGKF 572
Query: 179 GIICIEDLIHEILTVGPHFREANNFLWPFKLKAPLGGMKKKRNHYVEGGDAGNREDYINE 238
GI+CIED++H+I GPHF+E F+WPF+L P G+K + + EGGD GNRED INE
Sbjct: 573 GIVCIEDMVHQIYNAGPHFKEVARFMWPFELNKPAEGLKGSKTIFKEGGDTGNREDLINE 752
Query: 239 LIRRMN 244
LI +MN
Sbjct: 753 LINKMN 770
>AW397609 similar to GP|11994560|db 60S ribosomal protein L7 {Arabidopsis
thaliana}, partial (66%)
Length = 556
Score = 221 bits (562), Expect = 3e-58
Identities = 120/182 (65%), Positives = 139/182 (75%)
Frame = +2
Query: 9 VVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQKELIALK 68
+VP SVL +K EEWAL KQE ++A K R+ +R L + R+ A +YD +LI LK
Sbjct: 11 LVPYSVLNNRKMEEEWALAFKQELDAANKIRAVSRHLFYCRSIPVANDYDLQ*MDLIRLK 190
Query: 69 REAKLKGGFYVDPEAKLLFIIRIRGINAMDPKSRKILQLLRLRQICNGVFLKVNKATVNM 128
REA L GGFYVDP+A+LLFIIRI GINA+DPKSRKIL LLRLRQI NGVFLKV +TVNM
Sbjct: 191 REAWL*GGFYVDPDAELLFIIRIGGINALDPKSRKILHLLRLRQIFNGVFLKVITSTVNM 370
Query: 129 LHRVEPYVTYGYPNLKSVRELIYKRGYGKVDRQRIALTDNSIIEQVLGKHGIICIEDLIH 188
LH V+PYVTYGYP+LKSVRELIYKR YG + +Q ALTDNSIIE GK II +DLIH
Sbjct: 371 LHIVDPYVTYGYPHLKSVRELIYKRWYG*LMKQTTALTDNSIIEHAPGKCRIISTDDLIH 550
Query: 189 EI 190
EI
Sbjct: 551 EI 556
>AW568174 similar to GP|11994560|db 60S ribosomal protein L7 {Arabidopsis
thaliana}, partial (55%)
Length = 424
Score = 197 bits (500), Expect = 5e-51
Identities = 102/139 (73%), Positives = 115/139 (82%)
Frame = +2
Query: 5 EVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQKEL 64
E K VPESV+KK+ RNEEWAL KK E ++AK+KR ETRKLI+ AKQY+KEYD+ QKEL
Sbjct: 8 EAKA*VPESVVKKENRNEEWALEKKHELDAAKRKRFETRKLIFDIAKQYSKEYDEQQKEL 187
Query: 65 IALKREAKLKGGFYVDPEAKLLFIIRIRGINAMDPKSRKILQLLRLRQICNGVFLKVNKA 124
I +REAKL GG+YV+PEAKLL IIR RGINAMDP + KILQL+RLRQI NGV LKVN A
Sbjct: 188 IR*RREAKLTGGYYVEPEAKLL*IIRRRGINAMDPSTTKILQLMRLRQINNGVSLKVNGA 367
Query: 125 TVNMLHRVEPYVTYGYPNL 143
TVNMLH VEPYVTYGY L
Sbjct: 368 TVNMLHMVEPYVTYGYTYL 424
>TC227263 weakly similar to UP|RL7_DICDI (P11874) 60S ribosomal protein L7,
partial (43%)
Length = 543
Score = 186 bits (473), Expect = 6e-48
Identities = 85/159 (53%), Positives = 118/159 (73%)
Frame = +3
Query: 86 LFIIRIRGINAMDPKSRKILQLLRLRQICNGVFLKVNKATVNMLHRVEPYVTYGYPNLKS 145
L IRI+G M P++RK+L L LR+ + VF+K ++ + L RVEPYVTYGYPNLKS
Sbjct: 18 LLNIRIQGKKDMHPRTRKVLYSLGLRRRFSAVFVKPSEGIMAKLQRVEPYVTYGYPNLKS 197
Query: 146 VRELIYKRGYGKVDRQRIALTDNSIIEQVLGKHGIICIEDLIHEILTVGPHFREANNFLW 205
++ELIYK+G+ +++++++ LTDN+IIEQ LGK GI+CIED++H+I GPHF+E F+W
Sbjct: 198 IKELIYKKGHARMEQRKVPLTDNNIIEQELGKFGIVCIEDMVHQIYNAGPHFKEVVRFMW 377
Query: 206 PFKLKAPLGGMKKKRNHYVEGGDAGNREDYINELIRRMN 244
PF+L P G+K + + E GD GNRED INELI +MN
Sbjct: 378 PFELNKPAEGLKGSKTSFKEDGDTGNREDLINELINKMN 494
>AW309988 weakly similar to GP|6687301|emb| 60S ribosomal protein L7
{Cyanophora paradoxa}, partial (31%)
Length = 337
Score = 101 bits (252), Expect = 3e-22
Identities = 48/109 (44%), Positives = 69/109 (63%)
Frame = -1
Query: 125 TVNMLHRVEPYVTYGYPNLKSVRELIYKRGYGKVDRQRIALTDNSIIEQVLGKHGIICIE 184
T+ L RV PYVTYGY +L S+ EL + + ++ ++++ L DN+IIEQ LG GI+CI+
Sbjct: 337 TMAKLQRVIPYVTYGYRSLTSI*ELFSTQAHARMGQRKVPLADNNIIEQELG*FGIVCIQ 158
Query: 185 DLIHEILTVGPHFREANNFLWPFKLKAPLGGMKKKRNHYVEGGDAGNRE 233
D++H I GPHF+E F+WPF+L P G K R + + G RE
Sbjct: 157 DMVHHIYKAGPHFKEVVTFMWPFELYKPAAGFKGSRPSFKKMLAPGFRE 11
>CA802337
Length = 381
Score = 32.0 bits (71), Expect = 0.26
Identities = 18/64 (28%), Positives = 37/64 (57%)
Frame = +3
Query: 14 VLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQKELIALKREAKL 73
+LKK+K+ ++ KK++++ KKK+ + +K + K+ K+ +K+ K++ K
Sbjct: 36 LLKKKKKKKKKKKKKKKKKKKKKKKKKKKKK----KKKKKKKKKKKKKKKKKKKKKKKKX 203
Query: 74 KGGF 77
GGF
Sbjct: 204GGGF 215
Score = 28.9 bits (63), Expect = 2.2
Identities = 16/61 (26%), Positives = 33/61 (53%)
Frame = +3
Query: 16 KKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQKELIALKREAKLKG 75
KK+K+ ++ KK++++ KKK+ + +K + K+ K+ +K K++ L G
Sbjct: 60 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKXGGGFKKKKFLXG 239
Query: 76 G 76
G
Sbjct: 240 G 242
Score = 28.1 bits (61), Expect = 3.8
Identities = 16/61 (26%), Positives = 34/61 (55%)
Frame = +1
Query: 16 KKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQKELIALKREAKLKG 75
KK+K+ ++ KK++++ KKK+ + +K + K+ K+ +K LK+++ G
Sbjct: 61 KKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKXGGGLKKKSF*XG 240
Query: 76 G 76
G
Sbjct: 241 G 243
>TC227653 similar to UP|O80383 (O80383) 98b, partial (78%)
Length = 1654
Score = 31.2 bits (69), Expect = 0.45
Identities = 17/51 (33%), Positives = 28/51 (54%)
Frame = +1
Query: 1 MAKEEVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAK 51
MAK+ + E KQK++EE A K+E+E K ++ E +L+ + K
Sbjct: 997 MAKKNKEQYALEMEAYKQKKDEEAAHFMKEEEEQMKLQKQEALQLLKKKEK 1149
>TC206495 weakly similar to GB|AAO42927.1|28416793|BT004681 At5g24660
{Arabidopsis thaliana;} , partial (59%)
Length = 623
Score = 28.5 bits (62), Expect = 2.9
Identities = 34/128 (26%), Positives = 60/128 (46%), Gaps = 2/128 (1%)
Frame = +1
Query: 3 KEEVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKL--IWSRAKQYAKEYDDS 60
++++ TVV ES LK KRNEE ++K+ +ES +++ R+L W R + + +
Sbjct: 139 EKKMPTVVAESELK--KRNEE---LEKELRESKEREEQMKRELQSAWERLRVAEEAEERL 303
Query: 61 QKELIALKREAKLKGGFYVDPEAKLLFIIRIRGINAMDPKSRKILQLLRLRQICNGVFLK 120
+L L+ EA + Y R ++ MD SR Q L L+ + + L
Sbjct: 304 CSQLGELEAEAVYQARDY-----------HARIVSLMDQLSR--AQSLLLKTSASSISLP 444
Query: 121 VNKATVNM 128
+ +N+
Sbjct: 445 SSS*IINI 468
>TC215112
Length = 1736
Score = 28.5 bits (62), Expect = 2.9
Identities = 13/56 (23%), Positives = 34/56 (60%)
Frame = -3
Query: 1 MAKEEVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKE 56
++ +++ ++ S LKK+K+ ++ KK++++ KKK+ + +K + K+ K+
Sbjct: 324 VSNRQIQCLIFFSCLKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKKK 157
>TC205993 weakly similar to UP|Q6ZC61 (Q6ZC61) Ribosomal protein L30p
family-like, partial (85%)
Length = 763
Score = 28.5 bits (62), Expect = 2.9
Identities = 19/52 (36%), Positives = 29/52 (55%), Gaps = 1/52 (1%)
Frame = +3
Query: 86 LFIIRIRGINAMDPKSRKILQLLRLRQICNGVFLKVNKATV-NMLHRVEPYV 136
++I +RGI R+ L+ LRLR+ CN ++ N TV ML +V+ V
Sbjct: 195 IYITLVRGIPGTRRLHRRTLEALRLRK-CNRTVMRWNTPTVRGMLQQVKRLV 347
>BU927326
Length = 445
Score = 28.1 bits (61), Expect = 3.8
Identities = 20/61 (32%), Positives = 36/61 (58%)
Frame = +2
Query: 26 LVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQKELIALKREAKLKGGFYVDPEAKL 85
L K+ Q+ K++++ R+LI + AKE + + E+ ALKRE+ +K V+ E ++
Sbjct: 173 LAKRYMQDYEKERKA--RELIEEICDELAKEIGEDKAEIEALKRES-MKLREEVEEERRM 343
Query: 86 L 86
L
Sbjct: 344 L 346
>TC232314 weakly similar to UP|Q6NLY8 (Q6NLY8) At4g36720, partial (50%)
Length = 780
Score = 28.1 bits (61), Expect = 3.8
Identities = 11/29 (37%), Positives = 20/29 (68%)
Frame = -3
Query: 21 NEEWALVKKQEQESAKKKRSETRKLIWSR 49
++EW V KQE +++ R E+RK+I+ +
Sbjct: 319 HQEWEAVAKQEMQTSCDNRVESRKVIFQQ 233
>BF069321
Length = 422
Score = 28.1 bits (61), Expect = 3.8
Identities = 11/29 (37%), Positives = 20/29 (68%)
Frame = -2
Query: 21 NEEWALVKKQEQESAKKKRSETRKLIWSR 49
++EW V KQE +++ R E+RK+I+ +
Sbjct: 190 HQEWEAVAKQEMQTSCDNRVESRKVIFQQ 104
>TC219295
Length = 646
Score = 27.7 bits (60), Expect = 4.9
Identities = 20/62 (32%), Positives = 32/62 (51%)
Frame = +2
Query: 141 PNLKSVRELIYKRGYGKVDRQRIALTDNSIIEQVLGKHGIICIEDLIHEILTVGPHFREA 200
PNLKS+ K G G + R ++ +T + + V + I +ED++ E+ TVG F
Sbjct: 353 PNLKSL-----KTG-GGIKRIKVIITKQQLQKLVTKQ---ISVEDILSEVQTVGVKFSSN 505
Query: 201 NN 202
N
Sbjct: 506 QN 511
>TC217199 weakly similar to UP|Q7X9D9 (Q7X9D9) ABRH1 (Fragment), partial
(49%)
Length = 1061
Score = 26.9 bits (58), Expect = 8.4
Identities = 18/69 (26%), Positives = 34/69 (49%), Gaps = 5/69 (7%)
Frame = +2
Query: 11 PESVLKKQKRNEEWALVKKQEQESAKK-----KRSETRKLIWSRAKQYAKEYDDSQKELI 65
P ++L +N++ LV++ + S +K + K+ + AK + DD+++E
Sbjct: 260 PANILPTFDQNDKRTLVQEFQMSSIQKDFCFDELESIVKIKQAEAKMFQSRADDARREAE 439
Query: 66 ALKREAKLK 74
LKR A K
Sbjct: 440 GLKRIALAK 466
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.138 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,225,888
Number of Sequences: 63676
Number of extensions: 111647
Number of successful extensions: 700
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 684
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 695
length of query: 244
length of database: 12,639,632
effective HSP length: 95
effective length of query: 149
effective length of database: 6,590,412
effective search space: 981971388
effective search space used: 981971388
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC148819.1


