

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148775.5 + phase: 0 /pseudo
(302 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
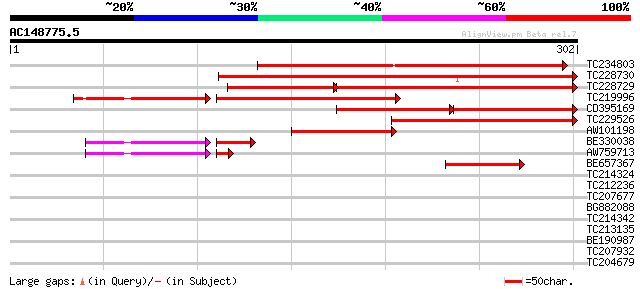
TC234803 similar to GB|AAQ62442.1|34146868|BT010441 At3g47590 {A... 256 7e-69
TC228730 229 2e-60
TC228729 157 4e-58
TC219996 weakly similar to GB|AAQ62442.1|34146868|BT010441 At3g4... 159 6e-52
CD395169 102 1e-44
TC229526 similar to UP|Q9FXF9 (Q9FXF9) F1N18.12 protein (At1g298... 157 6e-39
AW101198 82 4e-16
BE330038 weakly similar to GP|13161357|dbj P0013G02.26 {Oryza sa... 51 7e-14
AW759713 62 2e-10
BE657367 weakly similar to GP|19699292|gb AT4g24140/T19F6_130 {A... 45 4e-05
TC214324 weakly similar to UP|Q59538 (Q59538) Lipase-esterase ,... 36 0.019
TC212236 similar to UP|Q8LJ04 (Q8LJ04) OJ1116_H09.22 protein, pa... 31 0.60
TC207677 29 2.3
BG882088 29 3.0
TC214342 similar to UP|Q7TRQ1 (Q7TRQ1) Olfactory receptor Olfr64... 29 3.0
TC213135 similar to UP|Q9LJZ9 (Q9LJZ9) Protein arginine N-methyl... 28 5.1
BE190987 28 6.7
TC207932 similar to UP|Q7XYX6 (Q7XYX6) AKIN beta3, partial (91%) 28 6.7
TC204679 homologue to UP|Q8GZR6 (Q8GZR6) GcpE, partial (42%) 28 6.7
>TC234803 similar to GB|AAQ62442.1|34146868|BT010441 At3g47590 {Arabidopsis
thaliana;} , partial (45%)
Length = 557
Score = 256 bits (655), Expect = 7e-69
Identities = 126/165 (76%), Positives = 142/165 (85%)
Frame = +1
Query: 133 QHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVNLSGRYDLKAGIEERLGKDYLER 192
QHF +N + AI+GHSKGG VVLLYASK+H+IKTVVNLSGRYDLKAG+EERLGKDYLER
Sbjct: 1 QHFHGANHKVIAIIGHSKGGSVVLLYASKHHDIKTVVNLSGRYDLKAGLEERLGKDYLER 180
Query: 193 IRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACLQIDKDCRILTIHGSSDEIIPVQ 252
I KDGF DV +S G DYRVT ESLMDRL TNMHEACLQIDK+CR+LT+HGSSD +IPV
Sbjct: 181 IMKDGFIDVMQS-GSFDYRVTLESLMDRLDTNMHEACLQIDKECRVLTVHGSSDPVIPVG 357
Query: 253 DAHEFAKIIPNHKLHIIEGADHAYNNHQDELSSVFMSFIKETIDH 297
DA EFAKIIPNHKL IIEGADH+Y NHQDEL+SV ++ IKE + H
Sbjct: 358 DASEFAKIIPNHKLIIIEGADHSYTNHQDELASVVVNRIKEALVH 492
>TC228730
Length = 1544
Score = 229 bits (583), Expect = 2e-60
Identities = 120/230 (52%), Positives = 147/230 (63%), Gaps = 39/230 (16%)
Frame = +3
Query: 112 SEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVNL 171
SEGSF+YGNY++E +DL AV QHF E I+AIVGHSKGG+VVLLYASKY +I VVN+
Sbjct: 3 SEGSFQYGNYYREAEDLRAVVQHFCEQKYAIKAIVGHSKGGNVVLLYASKYKDIHIVVNI 182
Query: 172 SGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACLQ 231
SGR++L G+E RLGK +++RI++DG+ DVK GK+ YRVTEESLMDRL T H ACL
Sbjct: 183 SGRFNLARGMEGRLGKKFIQRIKQDGYIDVKNKRGKIMYRVTEESLMDRLSTITHLACLL 362
Query: 232 IDKDCR---------------------------------------ILTIHGSSDEIIPVQ 252
I +DCR +LTIHGS DEI+P +
Sbjct: 363 IPQDCRYSLLLHLITGIIENLAKSVASCLTVTTYHC*SLHQR*YSVLTIHGSMDEIVPAE 542
Query: 253 DAHEFAKIIPNHKLHIIEGADHAYNNHQDELSSVFMSFIKETIDHSKSTA 302
DA EF K I NH+L IEGADH Y HQDEL+S+ + FIK ID K T+
Sbjct: 543 DAVEFTKFISNHELCFIEGADHEYTYHQDELTSLVLEFIKIHIDKDKDTS 692
>TC228729
Length = 1249
Score = 157 bits (397), Expect(2) = 4e-58
Identities = 76/128 (59%), Positives = 94/128 (73%)
Frame = +1
Query: 175 YDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACLQIDK 234
+ L G+E RLGK +++RI++DG+ DVK GK+ YRVTEESLMDRL T H ACL I +
Sbjct: 175 FHLARGMEGRLGKKFIQRIKQDGYIDVKNKRGKIMYRVTEESLMDRLSTITHLACLLIPQ 354
Query: 235 DCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQDELSSVFMSFIKET 294
DC +LTIHGS DEI+P +DA EFAK I NH+L IEGADH Y HQDEL+S+ + FIK
Sbjct: 355 DCSVLTIHGSMDEIVPAEDALEFAKFISNHELCFIEGADHEYTYHQDELTSLVLEFIKIH 534
Query: 295 IDHSKSTA 302
ID K T+
Sbjct: 535 IDKDKDTS 558
Score = 85.5 bits (210), Expect(2) = 4e-58
Identities = 39/59 (66%), Positives = 48/59 (81%)
Frame = +2
Query: 117 EYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVNLSGRY 175
+YGNY++E +DL AV QHF E I+AIVGHSKGG+VVLLYASKY +I VVN+SGR+
Sbjct: 2 QYGNYYREAEDLRAVVQHFCEQKYAIKAIVGHSKGGNVVLLYASKYKDIHIVVNISGRF 178
>TC219996 weakly similar to GB|AAQ62442.1|34146868|BT010441 At3g47590
{Arabidopsis thaliana;} , partial (49%)
Length = 855
Score = 159 bits (402), Expect(2) = 6e-52
Identities = 76/98 (77%), Positives = 84/98 (85%)
Frame = +2
Query: 111 ESEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVN 170
ES+GSF+YG YW+E +DL AV QHF ESNR + AIVGHSKGG VVLLYASKYH+IKTVVN
Sbjct: 350 ESDGSFQYGYYWREAEDLRAVIQHFHESNRGVSAIVGHSKGGGVVLLYASKYHDIKTVVN 529
Query: 171 LSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKL 208
LSGRYDLK GIEERLGKD++ERIRKDGF DV RS L
Sbjct: 530 LSGRYDLKVGIEERLGKDHIERIRKDGFIDVTRSGNLL 643
Score = 62.8 bits (151), Expect(2) = 6e-52
Identities = 40/73 (54%), Positives = 45/73 (60%)
Frame = +1
Query: 35 FSF*ALSESDNTQQKW*KTCWHIT*IFWNNNQRHCYFVSWFSLL*RYQSHIEPCCCIGKG 94
FS *A ++SDNTQQ *KT +IT*I+ Q C V FSLL R Q E CCC GK
Sbjct: 142 FSC*A-TQSDNTQQAR*KTSGYIT*IW---KQGDCNLVPRFSLLKRIQFLSESCCCFGKC 309
Query: 95 TD*FFPF*LFWKW 107
+ FFP *L WKW
Sbjct: 310 QNEFFPL*LCWKW 348
>CD395169
Length = 608
Score = 102 bits (255), Expect(2) = 1e-44
Identities = 50/64 (78%), Positives = 53/64 (82%), Gaps = 1/64 (1%)
Frame = -1
Query: 175 YDLKAGIEERLGKDYLERIRKDGFFDVK-RSSGKLDYRVTEESLMDRLGTNMHEACLQID 233
YD+K GI E GKDYLERIRKDGF DVK R SG D+ VTEESLMDRLG NMHEACLQID
Sbjct: 602 YDMKPGIGEPFGKDYLERIRKDGFIDVKKRRSGSFDFHVTEESLMDRLGINMHEACLQID 423
Query: 234 KDCR 237
K+CR
Sbjct: 422 KECR 411
Score = 95.1 bits (235), Expect(2) = 1e-44
Identities = 44/66 (66%), Positives = 55/66 (82%)
Frame = -2
Query: 237 RILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQDELSSVFMSFIKETID 296
R+ TIHGSSDEIIPV+DA EFAKII NHKLH+IEGA+H ++NHQ +LSSV ++ I+ET+
Sbjct: 319 RVFTIHGSSDEIIPVEDAFEFAKIISNHKLHVIEGANHIFSNHQGDLSSVVVNCIEETLH 140
Query: 297 HSKSTA 302
K TA
Sbjct: 139 QVKGTA 122
>TC229526 similar to UP|Q9FXF9 (Q9FXF9) F1N18.12 protein (At1g29840), partial
(33%)
Length = 869
Score = 157 bits (397), Expect = 6e-39
Identities = 70/99 (70%), Positives = 87/99 (87%)
Frame = +3
Query: 204 SSGKLDYRVTEESLMDRLGTNMHEACLQIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPN 263
++G +YRVT ESLMDRL TNMHEACLQIDK+CR+LT+HGS D+++P DA+EFAKIIPN
Sbjct: 18 AAGNFEYRVTLESLMDRLDTNMHEACLQIDKECRVLTVHGSLDKVVPTDDAYEFAKIIPN 197
Query: 264 HKLHIIEGADHAYNNHQDELSSVFMSFIKETIDHSKSTA 302
HKLHIIEGADH++ NHQDEL+SV ++FIKET+ +STA
Sbjct: 198 HKLHIIEGADHSFTNHQDELASVVVNFIKETLHQDRSTA 314
>AW101198
Length = 394
Score = 81.6 bits (200), Expect = 4e-16
Identities = 36/56 (64%), Positives = 45/56 (80%)
Frame = +2
Query: 151 GGDVVLLYASKYHEIKTVVNLSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSG 206
G + LLYASKYH+IKT+VNLSG +DLK G+E R GKD+LER+RK+GF + K SG
Sbjct: 131 GANAALLYASKYHDIKTIVNLSGCHDLKVGLENRFGKDFLERLRKEGFIEFKAESG 298
>BE330038 weakly similar to GP|13161357|dbj P0013G02.26 {Oryza sativa
(japonica cultivar-group)}, partial (25%)
Length = 460
Score = 51.2 bits (121), Expect(2) = 7e-14
Identities = 29/67 (43%), Positives = 35/67 (51%)
Frame = +1
Query: 41 SESDNTQQKW*KTCWHIT*IFWNNNQRHCYFVSWFSLL*RYQSHIEPCCCIGKGTD*FFP 100
+ES N Q W T H *I+ NQR CY + W S+ * + H + CCC K FF
Sbjct: 202 AESHNN*QVWQ*TSGHFA*IW---NQRDCYPMPWSSIH*GRRYHKKSCCCSRKCRSQFFS 372
Query: 101 F*LFWKW 107
*L WKW
Sbjct: 373 L*LHWKW 393
Score = 43.1 bits (100), Expect(2) = 7e-14
Identities = 17/21 (80%), Positives = 20/21 (94%)
Frame = +2
Query: 111 ESEGSFEYGNYWKEVDDLHAV 131
ESEGSFE+G+YW+EVDDLH V
Sbjct: 395 ESEGSFEFGHYWREVDDLHDV 457
>AW759713
Length = 415
Score = 62.0 bits (149), Expect(2) = 2e-10
Identities = 36/67 (53%), Positives = 40/67 (58%)
Frame = +3
Query: 41 SESDNTQQKW*KTCWHIT*IFWNNNQRHCYFVSWFSLL*RYQSHIEPCCCIGKGTD*FFP 100
++SDNTQQ *KT +IT*I+ Q C V FSLL R Q E CCC GK FFP
Sbjct: 192 TQSDNTQQAR*KTSGYIT*IW---KQGDCNLVPRFSLLKRIQFLSESCCCFGKCQSEFFP 362
Query: 101 F*LFWKW 107
*L WKW
Sbjct: 363 L*LCWKW 383
Score = 20.8 bits (42), Expect(2) = 2e-10
Identities = 7/9 (77%), Positives = 9/9 (99%)
Frame = +1
Query: 111 ESEGSFEYG 119
ES+GSF+YG
Sbjct: 385 ESDGSFQYG 411
>BE657367 weakly similar to GP|19699292|gb AT4g24140/T19F6_130 {Arabidopsis
thaliana}, partial (31%)
Length = 708
Score = 45.1 bits (105), Expect = 4e-05
Identities = 16/42 (38%), Positives = 29/42 (68%)
Frame = -3
Query: 233 DKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADH 274
++DC++ HG +DE+IPV+ ++E K IP ++ +I+ DH
Sbjct: 412 NRDCKVTIFHGKNDEVIPVECSYEVQKRIPRAQVRVIDNKDH 287
>TC214324 weakly similar to UP|Q59538 (Q59538) Lipase-esterase , partial
(9%)
Length = 786
Score = 36.2 bits (82), Expect = 0.019
Identities = 19/86 (22%), Positives = 37/86 (42%), Gaps = 1/86 (1%)
Frame = +3
Query: 214 EESLMDRLGTNMHEACLQIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGAD 273
E L +HE C +L + G +D I+P +A +++IP +I+
Sbjct: 222 ESKTKPSLSKRLHEI------SCPVLIVTGDTDRIVPSWNAERLSRVIPGASFEVIKQCG 383
Query: 274 H-AYNNHQDELSSVFMSFIKETIDHS 298
H + +E S+ +F++ + S
Sbjct: 384 HLPHEEKVEEFISIVENFLRRLVSDS 461
>TC212236 similar to UP|Q8LJ04 (Q8LJ04) OJ1116_H09.22 protein, partial (13%)
Length = 896
Score = 31.2 bits (69), Expect = 0.60
Identities = 16/36 (44%), Positives = 20/36 (55%), Gaps = 3/36 (8%)
Frame = +1
Query: 235 DCRILTIHGSSDEII---PVQDAHEFAKIIPNHKLH 267
DC I++ HGS EII D F ++PNH LH
Sbjct: 40 DCNIVSKHGSQSEIILRVKQGDNPTFGFLMPNHHLH 147
>TC207677
Length = 1373
Score = 29.3 bits (64), Expect = 2.3
Identities = 18/68 (26%), Positives = 32/68 (46%), Gaps = 1/68 (1%)
Frame = +1
Query: 236 CRILTIHGSSDEIIPVQDAHEFAKIIP-NHKLHIIEGADHAYNNHQDELSSVFMSFIKET 294
C +L IHG++DE++ V + ++ ++ + G H E F+ +T
Sbjct: 721 CPVLVIHGTADEVVDVSHGKQLWELCKVKYEPLWVSGGGHCNLELYPEFIKHLKKFV-QT 897
Query: 295 IDHSKSTA 302
I SK+TA
Sbjct: 898 IGKSKATA 921
>BG882088
Length = 421
Score = 28.9 bits (63), Expect = 3.0
Identities = 12/31 (38%), Positives = 15/31 (47%)
Frame = -2
Query: 61 FWNNNQRHCYFVSWFSLL*RYQSHIEPCCCI 91
FW NN H W+ *RY+ + C CI
Sbjct: 207 FWGNNLLHI----WYFFF*RYKESFKACICI 127
>TC214342 similar to UP|Q7TRQ1 (Q7TRQ1) Olfactory receptor Olfr645, partial
(6%)
Length = 596
Score = 28.9 bits (63), Expect = 3.0
Identities = 17/47 (36%), Positives = 23/47 (48%)
Frame = +2
Query: 8 PC*FLIFNINSSTK*KFMNDLLSLICFFSF*ALSESDNTQQKW*KTC 54
PC L +ST KF+ LS CFF + + + Q +W KTC
Sbjct: 371 PCNQLD*QHYASTSTKFLGCCLSFACFFKWRLIKTAC*LQFEWCKTC 511
>TC213135 similar to UP|Q9LJZ9 (Q9LJZ9) Protein arginine
N-methyltransferase-like protein, partial (24%)
Length = 650
Score = 28.1 bits (61), Expect = 5.1
Identities = 15/35 (42%), Positives = 21/35 (59%)
Frame = -1
Query: 11 FLIFNINSSTK*KFMNDLLSLICFFSF*ALSESDN 45
F+ FNI +S ++ ND+ +ICF F LS S N
Sbjct: 218 FINFNILNSPM-QYNNDIRQVICFHDFICLSHSKN 117
>BE190987
Length = 544
Score = 27.7 bits (60), Expect = 6.7
Identities = 9/17 (52%), Positives = 12/17 (69%)
Frame = -1
Query: 105 WKWPSRESEGSFEYGNY 121
W+W +EG F+YGNY
Sbjct: 382 WRWSCLVTEGWFKYGNY 332
>TC207932 similar to UP|Q7XYX6 (Q7XYX6) AKIN beta3, partial (91%)
Length = 752
Score = 27.7 bits (60), Expect = 6.7
Identities = 13/35 (37%), Positives = 17/35 (48%), Gaps = 4/35 (11%)
Frame = -3
Query: 61 FWNNNQRHCYFVSWFSLL*RYQSHIEP----CCCI 91
FWN RH YF+ F + + +EP C CI
Sbjct: 282 FWNFKSRHSYFIMIFRVRIAHLGELEPNR*RCECI 178
>TC204679 homologue to UP|Q8GZR6 (Q8GZR6) GcpE, partial (42%)
Length = 943
Score = 27.7 bits (60), Expect = 6.7
Identities = 26/85 (30%), Positives = 40/85 (46%), Gaps = 16/85 (18%)
Frame = +2
Query: 192 RIRKDGFFDVKRSSGKLDYRVTEESL------MDRLGTNMHEACLQIDKDCRILTIHGS- 244
R+ + F D + L+Y TEE ++++ T + E C + + RI T HGS
Sbjct: 224 RVNPENFADRRAQFETLEY--TEEDYQKELEHIEKVFTPLVEKCKKYGRAMRIGTNHGSL 397
Query: 245 SDEIIP---------VQDAHEFAKI 260
SD I+ V+ A EFA+I
Sbjct: 398 SDRIMSYYGDSPRGMVESAFEFARI 472
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.337 0.147 0.492
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,555,499
Number of Sequences: 63676
Number of extensions: 261861
Number of successful extensions: 1808
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 1793
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1804
length of query: 302
length of database: 12,639,632
effective HSP length: 97
effective length of query: 205
effective length of database: 6,463,060
effective search space: 1324927300
effective search space used: 1324927300
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148775.5


