

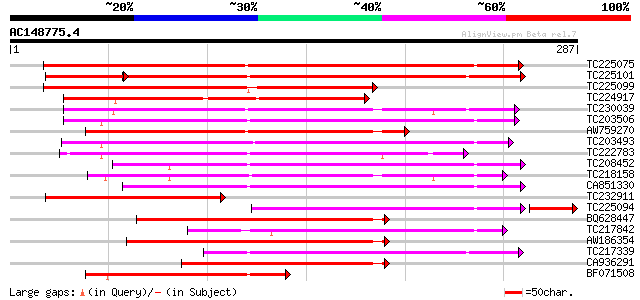
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148775.4 - phase: 0
(287 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC225075 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 224 3e-59
TC225101 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 180 2e-54
TC225099 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 174 4e-44
TC224917 similar to GB|AAK32841.1|13605696|AF361829 At2g45220/F4... 160 8e-40
TC230039 homologue to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase i... 144 4e-35
TC203506 similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (... 143 1e-34
AW759270 weakly similar to PIR|T49241|T49 pectinesterase-like pr... 140 5e-34
TC203493 similar to UP|Q9M3B0 (Q9M3B0) Pectinesterase-like prote... 137 7e-33
TC222783 weakly similar to UP|Q9SMV9 (Q9SMV9) Pectin methylester... 133 8e-32
TC208452 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (... 131 3e-31
TC218158 similar to UP|Q7XAF8 (Q7XAF8) Papillar cell-specific pe... 121 3e-28
CA851330 similar to PIR|T12995|T12 pectinesterase homolog T21L8.... 121 4e-28
TC232911 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 117 5e-27
TC225094 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B p... 113 8e-27
BQ628447 similar to GP|10178067|db pectin methylesterase-like pr... 114 5e-26
TC217842 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 prec... 110 7e-25
AW186354 110 7e-25
TC217339 homologue to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase p... 101 3e-22
CA936291 89 3e-18
BF071508 weakly similar to GP|10178035|dbj pectinesterase {Arabi... 86 1e-17
>TC225075 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (87%)
Length = 1744
Score = 224 bits (572), Expect = 3e-59
Identities = 127/244 (52%), Positives = 158/244 (64%), Gaps = 1/244 (0%)
Frame = +1
Query: 18 SSDILDTITQKGKFPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSK 77
+ D D + KG+FPSW+KP +RKLLQA AV D VA DGSGNY KIMDAV+AAP+ S
Sbjct: 517 AQDQFDAASSKGQFPSWIKPKERKLLQAIAVTPDVTVALDGSGNYAKIMDAVLAAPDYSM 696
Query: 78 KRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEG 137
KR+VI +KKGVY E+V I K N+M++G GM ATVI+G+ S D T + TF V G
Sbjct: 697 KRFVILVKKGVYVENVEIKKKKWNIMILGQGMDATVISGNRS-VVDGWTTFRSATFAVSG 873
Query: 138 LGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSE 197
GF A+DISF+NTA PE HQAVAL SDSD SVF+RC I G+QDSL + R
Sbjct: 874 RGFIARDISFQNTAGPEKHQAVALRSDSDLSVFFRCGIFGYQDSLYTHTMRQFFRDCTIS 1053
Query: 198 ARLTSYLVRQLSSFKTDIL-VRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEF 256
+ + F+ L V+KG Q+NTITA G + PN P GF+FQFCN+ AD +
Sbjct: 1054GTVDYIFGDATAVFQNCFLRVKKGLPNQKNTITAHGRKD-PNEPTGFSFQFCNITADSDL 1230
Query: 257 LPFV 260
+P V
Sbjct: 1231IPSV 1242
>TC225101 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (64%)
Length = 1177
Score = 180 bits (456), Expect(2) = 2e-54
Identities = 103/205 (50%), Positives = 131/205 (63%), Gaps = 1/205 (0%)
Frame = +3
Query: 58 GSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGD 117
G NY K+MDAV+AAPN S +RYVIHIK+GVY E+V I K NLMM+GDGM AT+I+G+
Sbjct: 207 GRENYTKVMDAVLAAPNYSMQRYVIHIKRGVYYENVEIKKKKWNLMMVGDGMDATIISGN 386
Query: 118 LSWGRDKLDTSYTYTFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISG 177
S+ D T + TF V G GF A+DI+F+NTA PE HQAVAL SDSD SVF+RC I G
Sbjct: 387 RSF-IDGWTTFRSATFAVSGRGFIARDITFQNTAGPEKHQAVALRSDSDLSVFFRCGIFG 563
Query: 178 FQDSLCANIKHHSIRIAKSEARLTSYLVRQLSSFKT-DILVRKGPTGQQNTITAQGGPEK 236
+QDSL + R K + + F+ I +KG Q+NTITA G +
Sbjct: 564 YQDSLYTHTMRQFYRECKISGTVDFIFGDATAIFQNCHISAKKGLPNQKNTITAH-GRKN 740
Query: 237 PNLPFGFAFQFCNVCADPEFLPFVN 261
P+ P GF+ QFCN+ AD + + VN
Sbjct: 741 PDEPTGFSIQFCNISADYDLVNSVN 815
Score = 50.4 bits (119), Expect(2) = 2e-54
Identities = 23/42 (54%), Positives = 31/42 (73%)
Frame = +2
Query: 19 SDILDTITQKGKFPSWVKPGDRKLLQASAVPADAVVASDGSG 60
SD + +G++PSWVK G+RKLLQA+ V DAVV +DG+G
Sbjct: 89 SDHFSFSSPQGQYPSWVKTGERKLLQANVVSFDAVVTADGTG 214
>TC225099 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (31%)
Length = 652
Score = 174 bits (441), Expect = 4e-44
Identities = 94/174 (54%), Positives = 119/174 (68%), Gaps = 5/174 (2%)
Frame = +2
Query: 18 SSDILDTITQKGKFPSWVKPGDRKLLQASAVPA-DAVVASDGSGNYMKIMDAVMAAPNGS 76
+ D D + KG+FP WVKP +RKLLQ+ + A D VA DGSGNY KIMDAV+AAP+ S
Sbjct: 122 AQDQFDDASSKGQFPLWVKPQERKLLQSIGMTAADVTVALDGSGNYAKIMDAVLAAPDYS 301
Query: 77 KKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLS----WGRDKLDTSYTYT 132
KR+VI +KKGVY E+V I K N+MM+G+GM + +I+G+ S W T + T
Sbjct: 302 MKRFVILVKKGVYVENVEIKRKKWNIMMVGEGMDSIIISGNWSDVYGW-----TTFRSAT 466
Query: 133 FGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANI 186
F V G+GF A+DI F NT WPE HQA+AL SD+D SVFYRCEI G D + A+I
Sbjct: 467 FAVSGIGFIARDIYFHNTEWPEKHQAMALRSDTDLSVFYRCEIVGDLDRIYADI 628
>TC224917 similar to GB|AAK32841.1|13605696|AF361829 At2g45220/F4L23.27
{Arabidopsis thaliana;} , partial (69%)
Length = 1483
Score = 160 bits (404), Expect = 8e-40
Identities = 88/157 (56%), Positives = 106/157 (67%), Gaps = 2/157 (1%)
Frame = +2
Query: 28 KGKFPSWVKPGDRKLLQASAVPADA--VVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIK 85
K FP+WVKPGDRKLLQ+S+V ++A VVA DGSG Y + AV AAP S RYVI++K
Sbjct: 662 KEGFPTWVKPGDRKLLQSSSVASNANVVVAKDGSGKYTTVKAAVDAAPKSSSGRYVIYVK 841
Query: 86 KGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDI 145
GVYNE V + + N+M++GDG+G T+ITG S G T + T G GF AQDI
Sbjct: 842 SGVYNEQVEVKGN--NIMLVGDGIGKTIITGSKSVGGGTT-TFRSATVAAVGDGFIAQDI 1012
Query: 146 SFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSL 182
+FRNTA NHQAVA S SD SVFYRC GFQD+L
Sbjct: 1013TFRNTAGAANHQAVAFRSGSDLSVFYRCSFEGFQDTL 1123
>TC230039 homologue to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isoform alpha
(Fragment) , partial (70%)
Length = 913
Score = 144 bits (364), Expect = 4e-35
Identities = 93/238 (39%), Positives = 127/238 (53%), Gaps = 7/238 (2%)
Frame = +1
Query: 28 KGKFPSWVKPGDRKLLQASAVPAD--AVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIK 85
K FP+W+ DRKLLQA+ + +VA DG+GN+ I +AV APN S R+VIHIK
Sbjct: 151 KDGFPTWLSTKDRKLLQAAVNETNFNLLVAKDGTGNFTTIAEAVAVAPNSSATRFVIHIK 330
Query: 86 KGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDI 145
G Y E+V + K+NLM +GDG+G TV+ + D T + T V G GF A+ I
Sbjct: 331 AGAYFENVEVIRKKTNLMFVGDGIGKTVVKASRN-VVDGWTTFQSATVAVVGDGFIAKGI 507
Query: 146 SFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLV 205
+F N+A P HQAVAL S SD S FY+C +QD+L HS+R + + +
Sbjct: 508 TFENSAGPSKHQAVALRSGSDFSAFYKCSFVAYQDTLYV----HSLRQFYRDCDVYGTVD 675
Query: 206 RQLSSFKT-----DILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLP 258
+ T ++ RK Q+N TAQ G E PN G + C V A + +P
Sbjct: 676 FIFGNAATVLQNCNLYARKPNENQRNLFTAQ-GREDPNQNTGISILNCKVAAAADLIP 846
>TC203506 similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (57%)
Length = 1320
Score = 143 bits (360), Expect = 1e-34
Identities = 87/235 (37%), Positives = 124/235 (52%), Gaps = 4/235 (1%)
Frame = +2
Query: 28 KGKFPSWVKPGDRKLLQA--SAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIK 85
+GKFP W+ DR+LL SA+ AD V+ G+G I +A+ AP S +R++I+++
Sbjct: 2 EGKFPGWLNKRDRRLLSLPLSAIQADITVSKTGNGTVKTIAEAIKKAPEHSTRRFIIYVR 181
Query: 86 KGVYNEH-VMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQD 144
G Y E+ + + K+N+M IGDG G TVITG + D + T ++ +F G GF A+D
Sbjct: 182 AGRYEENNLKVGRKKTNVMFIGDGKGKTVITGKKNV-IDGMTTFHSASFAASGAGFIARD 358
Query: 145 ISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYL 204
I+F N A P HQAVAL +D +V YRC I G+QDS + R +
Sbjct: 359 ITFENYAGPAKHQAVALRVGADHAVVYRCNIVGYQDSCYVHSNRQFFRECNIYGTVDFIF 538
Query: 205 VRQLSSF-KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLP 258
F K +I RK Q+NTITAQ + PN G + C + P+ P
Sbjct: 539 GNAAVVFQKCNIYARKPMAQQKNTITAQNRKD-PNQNTGISLHNCRILPAPDLAP 700
>AW759270 weakly similar to PIR|T49241|T49 pectinesterase-like protein -
Arabidopsis thaliana, partial (33%)
Length = 586
Score = 140 bits (354), Expect = 5e-34
Identities = 85/165 (51%), Positives = 109/165 (65%), Gaps = 1/165 (0%)
Frame = +2
Query: 39 DRKLLQASAVPA-DAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVYNEHVMINN 97
++KLLQ+ + A D VA DGSGNY KIMDAV+AAP+ + KR I +KKGVY E+V I
Sbjct: 5 EKKLLQSIGMTAADVTVA*DGSGNYAKIMDAVLAAPDYNMKRIEILVKKGVYVENVEIKR 184
Query: 98 SKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRNTAWPENHQ 157
K N+MM G+GM +T+I G+ S D+ T T TF V G G+ A+DISF+NTA PE HQ
Sbjct: 185 KKWNIMMDGEGMDSTIIYGNRS-VNDRWTTFRTATFAVSGRGYIARDISFQNTARPEKHQ 361
Query: 158 AVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTS 202
AVAL SD+D SV +RC I QDSL H++R E +T+
Sbjct: 362 AVALRSDTDLSVIFRCGIFCDQDSLYT----HTMRQYFRECTITA 484
>TC203493 similar to UP|Q9M3B0 (Q9M3B0) Pectinesterase-like protein
(At3g49220), partial (59%)
Length = 1298
Score = 137 bits (344), Expect = 7e-33
Identities = 84/233 (36%), Positives = 125/233 (53%), Gaps = 4/233 (1%)
Frame = +1
Query: 27 QKGKFPSWVKPGDRKLLQA--SAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHI 84
++ FP+W+ DR+LL S + AD VV+ DG+G I +A+ P S +R +I+I
Sbjct: 127 REDNFPTWLNGRDRRLLSLPLSQIQADIVVSKDGNGTVKTIAEAIKKVPEYSSRRIIIYI 306
Query: 85 KKGVYNE-HVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQ 143
+ G Y E ++ + K+N+M IGDG G TVITG ++ ++ L T +T +F G GF A+
Sbjct: 307 RAGRYEEDNLKLGRKKTNVMFIGDGKGKTVITGGRNYYQN-LTTFHTASFAASGSGFIAK 483
Query: 144 DISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSY 203
D++F N A P HQAVAL +D +V YRC I G+QD++ + R +
Sbjct: 484 DMTFENYAGPGRHQAVALRVGADHAVVYRCNIIGYQDTMYVHSNRQFYRECDIYGTVDFI 663
Query: 204 LVRQLSSFKTDILVRKGPTGQQ-NTITAQGGPEKPNLPFGFAFQFCNVCADPE 255
F+ L + P QQ NTITAQ + PN G + C + A P+
Sbjct: 664 FGNAAVVFQNCTLWARKPMAQQKNTITAQNRKD-PNQNTGISIHNCRIMATPD 819
>TC222783 weakly similar to UP|Q9SMV9 (Q9SMV9) Pectin methylesterase
precursor , partial (29%)
Length = 630
Score = 133 bits (335), Expect = 8e-32
Identities = 80/213 (37%), Positives = 120/213 (55%), Gaps = 6/213 (2%)
Frame = +3
Query: 26 TQKGKFPSWVKPGDRKLLQA---SAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVI 82
T++G +P+WV DRKLL AV A VA DGSG + ++DA+ + P + RY+I
Sbjct: 3 TREG-YPTWVSAADRKLLAQLNDGAVLPHATVAKDGSGQFTTVLDAINSYPKKHQGRYII 179
Query: 83 HIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSA 142
++K G+Y+E++ ++ K NL++ GDG T+ITG ++ + T T TF F A
Sbjct: 180 YVKAGIYDEYITVDKKKPNLLIYGDGPTKTIITGRKNF-HEGTKTMRTATFSTVAEDFMA 356
Query: 143 QDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIK---HHSIRIAKSEAR 199
+ I+F NTA E HQAVAL D SVF+ C + G+QD+L A+ + + I+ +
Sbjct: 357 KSIAFENTAGAEGHQAVALRVQGDRSVFFDCAMRGYQDTLYAHAHRQFYRNCEISGTIDF 536
Query: 200 LTSYLVRQLSSFKTDILVRKGPTGQQNTITAQG 232
+ Y + + K ILVRK QQN + A G
Sbjct: 537 IFGYSTTLIQNSK--ILVRKPMPNQQNIVVADG 629
>TC208452 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (56%)
Length = 1273
Score = 131 bits (330), Expect = 3e-31
Identities = 77/213 (36%), Positives = 120/213 (56%), Gaps = 4/213 (1%)
Frame = +1
Query: 53 VVASDGSGNYMKIMDAVMAAPNGSKKR---YVIHIKKGVYNEHVMINNSKSNLMMIGDGM 109
+V+ G NY I DA+ AAPN +K +++++++G+Y E+V+I K N++++GDG+
Sbjct: 295 IVSHYGIDNYTSIGDAIAAAPNNTKPEDGYFLVYVREGLYEEYVVIPKEKKNILLVGDGI 474
Query: 110 GATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSV 169
T+ITG+ S D T + TF V G F A D++FRNTA PE HQAVA+ +++D S
Sbjct: 475 NKTIITGNHSV-IDGWTTFNSSTFAVSGERFIAVDVTFRNTAGPEKHQAVAVRNNADLST 651
Query: 170 FYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQLSSFK-TDILVRKGPTGQQNTI 228
FYRC G+QD+L + R + + F+ I RK Q+N +
Sbjct: 652 FYRCSFEGYQDTLYVHSLRQFYRECEIYGTVDFIFGNAAVVFQGCKIYARKPLPNQKNAV 831
Query: 229 TAQGGPEKPNLPFGFAFQFCNVCADPEFLPFVN 261
TAQG + PN G + Q C++ A P+ + +N
Sbjct: 832 TAQGRTD-PNQNTGISIQNCSIDAAPDLVADLN 927
>TC218158 similar to UP|Q7XAF8 (Q7XAF8) Papillar cell-specific pectin
methylesterase-like protein, partial (59%)
Length = 1291
Score = 121 bits (304), Expect = 3e-28
Identities = 89/226 (39%), Positives = 119/226 (52%), Gaps = 13/226 (5%)
Frame = +3
Query: 40 RKLLQASA----VPADAV-VASDGSGNYMKIMDAVMAAPNGSKKR---YVIHIKKGVYNE 91
RKLLQA V D V V+ DGSGN+ I DA+ AAPN S ++I++ GVY E
Sbjct: 102 RKLLQAKVGDEVVVRDIVTVSQDGSGNFTTINDAIAAAPNKSVSTDGYFLIYVTAGVYEE 281
Query: 92 HVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRNTA 151
+V I+ K+ LMM+GDG+ T+ITG+ S D T + T V G GF +++ RNTA
Sbjct: 282 NVSIDKKKTYLMMVGDGINKTIITGNRSVV-DGWTTFSSATLAVVGQGFVGVNMTIRNTA 458
Query: 152 WPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQLSSF 211
HQAVAL S +D S FY C G+QD+L HS+R SE + + +
Sbjct: 459 GAVKHQAVALRSGADLSTFYSCSFEGYQDTLYV----HSLRQFYSECDIYGTVDFIFGNA 626
Query: 212 KT-----DILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCA 252
K + R +GQ N ITAQG + PN G + C + A
Sbjct: 627 KVVFQNCKMYPRLPMSGQFNAITAQGRTD-PNQDTGISIHNCTIRA 761
>CA851330 similar to PIR|T12995|T12 pectinesterase homolog T21L8.150 -
Arabidopsis thaliana, partial (37%)
Length = 700
Score = 121 bits (303), Expect = 4e-28
Identities = 79/206 (38%), Positives = 113/206 (54%), Gaps = 2/206 (0%)
Frame = +2
Query: 58 GSGNYMKIMDAVMAAPNGSKK-RYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITG 116
GSGN+ + DA+ AA +K R+VIH+KKGVY E++ + N+M++GDG+ T+IT
Sbjct: 14 GSGNFKTVQDALNAAAKRKEKTRFVIHVKKGVYRENIEVALHNDNIMLVGDGLRNTIITS 193
Query: 117 DLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEIS 176
S +D T + T G++GL F A+DI+F+N+A QAVAL S SD SVFYRC I
Sbjct: 194 ARSV-QDGYTTYSSATAGIDGLHFIARDITFQNSAGVHKGQAVALRSASDLSVFYRCGIM 370
Query: 177 GFQDSLCANIKHHSIRIAKSEARLTSYLVRQLSSFKT-DILVRKGPTGQQNTITAQGGPE 235
G+QD+L A+ + R + F+ I R+ GQ N ITAQG +
Sbjct: 371 GYQDTLMAHAQRQFYRQCYIYGTVDFIFGNAAVVFQNCYIFARRPLEGQANMITAQGRGD 550
Query: 236 KPNLPFGFAFQFCNVCADPEFLPFVN 261
P G + + A P+ P V+
Sbjct: 551 -PFQNTGISIHNSQIRAAPDLKPVVD 625
>TC232911 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (34%)
Length = 843
Score = 117 bits (294), Expect = 5e-27
Identities = 59/92 (64%), Positives = 72/92 (78%), Gaps = 1/92 (1%)
Frame = +2
Query: 19 SDILDTITQKGKFPSWVKPGDRKLLQAS-AVPADAVVASDGSGNYMKIMDAVMAAPNGSK 77
SD + +G FP WVKPG+RKLLQA+ V DAVVA+DG+GN+ K+MDAV+AAPN S
Sbjct: 560 SDHYTFSSPQGHFPPWVKPGERKLLQAANGVSFDAVVAADGTGNFTKVMDAVLAAPNYSM 739
Query: 78 KRYVIHIKKGVYNEHVMINNSKSNLMMIGDGM 109
+RYVIHIK+GVYNE+V I K NLMM+GDGM
Sbjct: 740 QRYVIHIKRGVYNENVEIKKKKWNLMMVGDGM 835
>TC225094 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (47%)
Length = 919
Score = 113 bits (283), Expect(2) = 8e-27
Identities = 65/140 (46%), Positives = 84/140 (59%), Gaps = 1/140 (0%)
Frame = +3
Query: 123 DKLDTSYTYTFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSL 182
D T + TF V G GF A+DISF+NTA PE HQAVAL SD+D SVF+RC I G+QDSL
Sbjct: 30 DGWTTFRSATFAVSGRGFIARDISFQNTAGPEKHQAVALRSDTDLSVFFRCGIFGYQDSL 209
Query: 183 CANIKHHSIRIAKSEARLTSYLVRQLSSFKTDIL-VRKGPTGQQNTITAQGGPEKPNLPF 241
+ R + + F+ L V+KG Q+NTITA G + PN P
Sbjct: 210 YTHTMRQFFRECTITGTVDYIFGDATAVFQNCFLRVKKGLPNQKNTITAHGRKD-PNEPT 386
Query: 242 GFAFQFCNVCADPEFLPFVN 261
GF+FQFCN+ AD + +P+V+
Sbjct: 387 GFSFQFCNITADSDLVPWVS 446
Score = 24.3 bits (51), Expect(2) = 8e-27
Identities = 10/24 (41%), Positives = 16/24 (66%)
Frame = +2
Query: 264 KHSSEDRRRLEALLTSSIHAVLNQ 287
+HS LE LLT+S HA++++
Sbjct: 446 QHSKLPGETLEELLTNSFHAIVHE 517
>BQ628447 similar to GP|10178067|db pectin methylesterase-like protein
{Arabidopsis thaliana}, partial (22%)
Length = 446
Score = 114 bits (285), Expect = 5e-26
Identities = 62/128 (48%), Positives = 83/128 (64%)
Frame = +2
Query: 65 IMDAVMAAPNGSKKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDK 124
+ +AV AAP+ +KR+VI+IK+GVY E V + K N++ +GDGMG TVITG + G+
Sbjct: 32 VQEAVNAAPDEGEKRFVIYIKEGVYEERVRVPLKKRNVVFLGDGMGKTVITGSANVGQPG 211
Query: 125 LDTSYTYTFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCA 184
+ T + T GV G GF A+D++ +NTA HQAVA SDSD SV CE G QD+L A
Sbjct: 212 MTTYNSATVGVAGDGFIAKDLTIQNTAGANAHQAVAFRSDSDLSVIENCEFIGNQDTLYA 391
Query: 185 NIKHHSIR 192
HS+R
Sbjct: 392 ----HSLR 403
>TC217842 similar to UP|PME2_CITSI (O04887) Pectinesterase 2 precursor
(Pectin methylesterase) (PE) , partial (48%)
Length = 1225
Score = 110 bits (275), Expect = 7e-25
Identities = 67/166 (40%), Positives = 94/166 (56%), Gaps = 4/166 (2%)
Frame = +3
Query: 91 EHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTY---TFGVEGLGFSAQDISF 147
E++ I + NLM++GDGMGAT++TG+ + +D S T+ TF V+G GF A+DI+F
Sbjct: 9 ENIDIKRTVKNLMIVGDGMGATIVTGN----HNAIDGSTTFRSATFAVDGDGFIARDITF 176
Query: 148 RNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQ 207
NTA P+ HQAVAL S +D SVFYRC G+QD+L R +
Sbjct: 177 ENTAGPQKHQAVALRSGADHSVFYRCSFRGYQDTLYVYANRQFYRDCDIYGTVDFIFGDA 356
Query: 208 LSSFKT-DILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCA 252
++ + +I VRK + QQNT+TAQG + PN G C + A
Sbjct: 357 VAVLQNCNIYVRKPMSNQQNTVTAQGRTD-PNENTGIIIHNCRITA 491
>AW186354
Length = 432
Score = 110 bits (275), Expect = 7e-25
Identities = 62/133 (46%), Positives = 83/133 (61%)
Frame = +3
Query: 60 GNYMKIMDAVMAAPNGSKKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLS 119
G Y + +AV AAP KR+VI+IK+GVY E V I K N++ +GDG+G TVITG+ +
Sbjct: 3 GCYKTVQEAVNAAPANGTKRFVIYIKEGVYEETVRIPLEKRNVVFLGDGIGKTVITGNGN 182
Query: 120 WGRDKLDTSYTYTFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQ 179
G+ + T + T V G GF A++++ NTA P+ HQAVA DSD SV CE G Q
Sbjct: 183 VGQQGMTTYNSATVAVLGDGFMAKELTVENTAGPDAHQAVAFRLDSDLSVIENCEFLGNQ 362
Query: 180 DSLCANIKHHSIR 192
D+L A HS+R
Sbjct: 363 DTLYA----HSLR 389
>TC217339 homologue to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase precursor,
partial (48%)
Length = 923
Score = 101 bits (252), Expect = 3e-22
Identities = 67/163 (41%), Positives = 87/163 (53%), Gaps = 1/163 (0%)
Frame = +1
Query: 99 KSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRNTAWPENHQA 158
K+N+M++GDG ATVITG+L++ D T T T G GF AQDI F+NTA P+ HQA
Sbjct: 4 KTNVMLVGDGKDATVITGNLNF-IDGTTTFKTATVAAVGDGFIAQDIWFQNTAGPQKHQA 180
Query: 159 VALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQLSSF-KTDILV 217
VAL +D SV RC I FQD+L A+ R + + F K D++
Sbjct: 181 VALRVGADQSVINRCRIDAFQDTLYAHSNRQFYRDSFITGTVDFIFGNAAVVFQKCDLVA 360
Query: 218 RKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLPFV 260
RK Q N +TAQ G E PN G + Q CN+ + P V
Sbjct: 361 RKPMDKQNNMVTAQ-GREDPNQNTGTSIQQCNLTPSSDLKPVV 486
>CA936291
Length = 429
Score = 88.6 bits (218), Expect = 3e-18
Identities = 49/105 (46%), Positives = 65/105 (61%)
Frame = +1
Query: 88 VYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISF 147
VY E V + +K N++ +GDG+G TVITGD + G+ + T + T V G GF A+D++
Sbjct: 1 VYQETVRVPLAKRNVVFLGDGIGKTVITGDANVGQQGMTTYNSATVAVLGDGFMAKDLTI 180
Query: 148 RNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIR 192
NTA P+ HQAVA DSD SV CE G QD+L A HS+R
Sbjct: 181 ENTAGPDAHQAVAFRLDSDLSVIENCEFLGNQDTLYA----HSLR 303
>BF071508 weakly similar to GP|10178035|dbj pectinesterase {Arabidopsis
thaliana}, partial (17%)
Length = 318
Score = 86.3 bits (212), Expect = 1e-17
Identities = 50/106 (47%), Positives = 66/106 (62%), Gaps = 2/106 (1%)
Frame = +3
Query: 39 DRKLLQASAV--PADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVYNEHVMIN 96
DRKLL + A VVA DGSG Y KI DA+ PN S KR VI++K+GVY E+V +
Sbjct: 3 DRKLLLTEDLREKAHIVVAKDGSGKYKKISDALKHVPNNSNKRTVIYVKRGVYYENVRVE 182
Query: 97 NSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSA 142
+K N+M+IGDGM +T+++G ++ D T T TF V G F A
Sbjct: 183 KTKWNVMIIGDGMTSTIVSGSRNF-VDGTPTFSTATFAVFGRNFIA 317
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,854,532
Number of Sequences: 63676
Number of extensions: 171348
Number of successful extensions: 1161
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 1110
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1114
length of query: 287
length of database: 12,639,632
effective HSP length: 96
effective length of query: 191
effective length of database: 6,526,736
effective search space: 1246606576
effective search space used: 1246606576
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148775.4


