

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148758.4 - phase: 0 /pseudo
(618 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
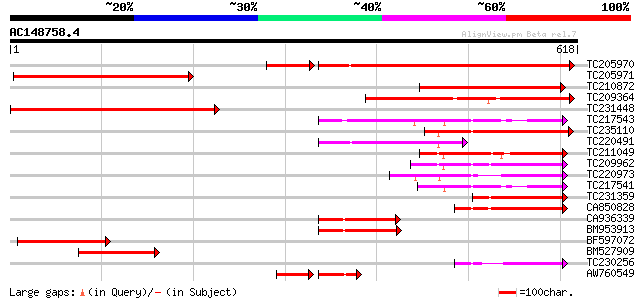
Score E
Sequences producing significant alignments: (bits) Value
TC205970 488 e-157
TC205971 330 1e-90
TC210872 228 5e-60
TC209364 215 4e-56
TC231448 204 7e-53
TC217543 similar to UP|Q940R0 (Q940R0) At1g29690/F15D2_24, parti... 201 8e-52
TC235110 similar to PIR|C86410|C86410 protein F3M18.18 [imported... 174 8e-44
TC220491 135 4e-32
TC211049 similar to UP|Q9LQV3 (Q9LQV3) F10B6.18, partial (19%) 131 8e-31
TC209962 128 7e-30
TC220973 113 3e-25
TC217541 110 2e-24
TC231359 106 3e-23
CA850828 weakly similar to GP|14209545|dbj P0481E12.26 {Oryza sa... 102 7e-22
CA936339 97 2e-20
BM953913 96 6e-20
BF597072 91 2e-18
BM527909 weakly similar to PIR|C86410|C864 protein F3M18.18 [imp... 87 2e-17
TC230256 73 4e-13
AW760549 57 1e-11
>TC205970
Length = 1317
Score = 488 bits (1257), Expect(2) = e-157
Identities = 238/279 (85%), Positives = 257/279 (91%)
Frame = +3
Query: 337 KPAIEELHQFLEFQLPRQWAPVFGELALGPDRKKSQSSSSLQFSFMGPKLYVNTSPVVVG 396
KP IEELHQFLEFQLPRQWAPVFGELALGP+RK Q+++SLQFSFMGPKLYVNT+PV VG
Sbjct: 162 KPQIEELHQFLEFQLPRQWAPVFGELALGPERKP-QNTASLQFSFMGPKLYVNTTPVDVG 338
Query: 397 MKPVTGLRLYLEGKKSNCLAIHLQHLSSLPKTFQLKDETNRNVSDASSERKYYEKVQWKS 456
KPVTGLRLYLEGK+SNCLAIHLQHLSSLPKTFQL+DE N N S+ SSERKYYEKVQWKS
Sbjct: 339 KKPVTGLRLYLEGKRSNCLAIHLQHLSSLPKTFQLQDEPNGNASNDSSERKYYEKVQWKS 518
Query: 457 FSHICTAPVESYDDNAVVTGAHFEVGETGLKKVLFLRLHFCKVADATRVRAPEWDGSPGL 516
FSH+CTAPV+S DDNAVVTGAHFEVG+TGLKKVLFLRLHFCKV ATRV+ PEW+GSPGL
Sbjct: 519 FSHVCTAPVDSDDDNAVVTGAHFEVGDTGLKKVLFLRLHFCKVVGATRVKVPEWEGSPGL 698
Query: 517 TQKSGMISTFISTRFSGPQKLPPPQPSDVNVNSALYPGGPPVPAQAPKLLKFVDTTEMTR 576
TQKSG+IST IST FSGPQK PPP+PSDVN+NSALYPGGPPVP Q+PKLL+FVDTTEMTR
Sbjct: 699 TQKSGIISTLISTTFSGPQKPPPPRPSDVNINSALYPGGPPVPTQSPKLLRFVDTTEMTR 878
Query: 577 GPQDLPGYWVVSGARLYVEKGKISLKVKYSLLTVILQDE 615
GPQD PGYWVVSGARL VEK KISLKVKYSLLTVI +E
Sbjct: 879 GPQDSPGYWVVSGARLLVEKAKISLKVKYSLLTVIPDEE 995
Score = 87.4 bits (215), Expect(2) = e-157
Identities = 44/52 (84%), Positives = 46/52 (87%)
Frame = +1
Query: 281 VEMGNKISATMSGAKLFCLNLM*YQCHSYQLHLYLVE*MGVDI*LMP*ISIY 332
VEMG + +SGAKLFCLNLM*YQCH YQLHLYLVE*MGVDI*LMP*ISIY
Sbjct: 1 VEMGKDLCPIVSGAKLFCLNLM*YQCHLYQLHLYLVE*MGVDI*LMP*ISIY 156
>TC205971
Length = 673
Score = 330 bits (846), Expect = 1e-90
Identities = 158/196 (80%), Positives = 180/196 (91%)
Frame = +1
Query: 5 KKVLSVEDVIKSVGLGYDLTNDLRLKFCKYDSKLIAIDHDNLRTVELPGRVSIPNVPKSI 64
KK ++ ED I+++GLGYDLTNDL+LKFCK S+LIAID DNLRTVELP R+SIPNVPKSI
Sbjct: 85 KKGVAAEDAIRAIGLGYDLTNDLKLKFCKNHSRLIAIDDDNLRTVELPPRISIPNVPKSI 264
Query: 65 NCDKGDRMRLCSDVLSFQQMSEQFNQEVSLSGKIPTGHFNSAFQFSGVWQRDAANTKSLA 124
CDKGDRMRLCSDVLSFQQMSEQFNQ++SLSGKIPTGHFN+AF F+GVWQ+DAANTK+LA
Sbjct: 265 KCDKGDRMRLCSDVLSFQQMSEQFNQDLSLSGKIPTGHFNTAFGFTGVWQKDAANTKTLA 444
Query: 125 FDGVSITLYNIALDKTHVVLSDHVKRAVPSSWDPAALARFIEKYGTHAVVGVKIGGTDII 184
FDGVSITLY+IA +KT VVL DHVK+AVPSSWDPAAL RFIEKYGTH +VGVK+GGTDII
Sbjct: 445 FDGVSITLYDIAFEKTQVVLHDHVKQAVPSSWDPAALTRFIEKYGTHVIVGVKMGGTDII 624
Query: 185 YAKQQYSSPLQPSDVQ 200
YAKQQYSS + P++VQ
Sbjct: 625 YAKQQYSSTVPPAEVQ 672
>TC210872
Length = 484
Score = 228 bits (582), Expect = 5e-60
Identities = 106/160 (66%), Positives = 130/160 (81%)
Frame = +3
Query: 447 KYYEKVQWKSFSHICTAPVESYDDNAVVTGAHFEVGETGLKKVLFLRLHFCKVADATRVR 506
++YEKVQWK+FSH+CTAPVES +D ++VTGA +V G+K +LFLRL F V A V+
Sbjct: 3 RFYEKVQWKNFSHVCTAPVESEEDLSIVTGAQLQVENYGIKNILFLRLRFSTVLGAKAVK 182
Query: 507 APEWDGSPGLTQKSGMISTFISTRFSGPQKLPPPQPSDVNVNSALYPGGPPVPAQAPKLL 566
PEW+GS L KSG+IST IS F+ + PPP+P+DVN+NSA+YPGGPPVP QAPKLL
Sbjct: 183 HPEWEGSLKLGAKSGLISTLISQHFTSTFQKPPPRPADVNINSAVYPGGPPVPVQAPKLL 362
Query: 567 KFVDTTEMTRGPQDLPGYWVVSGARLYVEKGKISLKVKYS 606
KFVDTTEMTRGPQ+ PGYWV+SGA+L V+KGKISL+VKYS
Sbjct: 363 KFVDTTEMTRGPQESPGYWVISGAKLVVDKGKISLRVKYS 482
>TC209364
Length = 725
Score = 215 bits (548), Expect = 4e-56
Identities = 118/231 (51%), Positives = 151/231 (65%), Gaps = 4/231 (1%)
Frame = +2
Query: 389 NTSPVVVGMKPVTGLRLYLEGKKSNCLAIHLQHLSSLPKTFQLKDETNRNVSDASSERKY 448
NT PV VG +PV GLRL LEG+ SN LAIHLQHL+SLPK+ + D +N +S S
Sbjct: 2 NTIPVDVGNRPVVGLRLQLEGRSSNRLAIHLQHLASLPKSLSVSDNSNAYLSCDSYNCNL 181
Query: 449 YEKVQWKSFSHICTAPVESYDDNAVVTGAHFEVGETGLKKVLFLRLHFCKVADATRVRAP 508
++KV+W S S++CTAPVES D ++VTGA +V K LFLRL F KV T +AP
Sbjct: 182 HKKVKWNSLSYVCTAPVESDDSVSIVTGAQLQVEN----KCLFLRLCFSKVIGVTLRKAP 349
Query: 509 EWDGSPGLTQKS----GMISTFISTRFSGPQKLPPPQPSDVNVNSALYPGGPPVPAQAPK 564
EWD SP L Q S G+++TFIS ++ P+P DV + S++Y P + PK
Sbjct: 350 EWDQSPSLGQFSIKGGGILTTFISKA----EQRDHPKPGDVTIGSSIYSAARLAPVRTPK 517
Query: 565 LLKFVDTTEMTRGPQDLPGYWVVSGARLYVEKGKISLKVKYSLLTVILQDE 615
LL+FVDTTE+ RGP D PG+WVVSGARL+V+ KI L VKYSL + +Q E
Sbjct: 518 LLRFVDTTEIMRGPVDTPGHWVVSGARLHVQDAKIYLHVKYSLFSFAMQTE 670
>TC231448
Length = 813
Score = 204 bits (520), Expect = 7e-53
Identities = 102/228 (44%), Positives = 152/228 (65%), Gaps = 1/228 (0%)
Frame = +3
Query: 2 SSKKKVLSVEDVIKSVGLGYDLTNDLRLKFCKYDSKLIAIDHDNLRTVELPGRVSIPNVP 61
+S ++ + ++ I S+GLG+D+T D+ CK S+LI ++ + R +E+PG VSIPNVP
Sbjct: 81 ASDLRLEAAQNAINSIGLGFDITQDISFDNCKKGSRLIFVNEEQCRHLEIPGGVSIPNVP 260
Query: 62 KSINCDKGDRMRLCSDVLSFQQMSEQFNQEVSLSGKIPTGHFNSAFQFSGVWQRDAANTK 121
SI C +G+ +R SDVL+ QM E FNQ++ LSG + +GH ++F S +D A+ K
Sbjct: 261 NSIKCVRGESIRFESDVLTRDQMMEHFNQQMLLSGNLASGHLCASFGLSDRSIKDLASIK 440
Query: 122 SLAFDGVSITLYNIALDKTHVVLSDHVKRAVPSSWDPAALARFIEKYGTHAVVGVKIGGT 181
SLA+DG I Y I L++ H + D V+ AVPSSWDP ALARFI+++GTH +VGV +GG
Sbjct: 441 SLAYDGWFIKRYTIELERHHCKILDQVEEAVPSSWDPEALARFIQRFGTHVIVGVSMGGK 620
Query: 182 DIIYAKQQYSSPLQPSDVQKKLKDMADELFRGQAGQNN-ANDGTFNSK 228
D++Y +Q+ +S L P+ +QK LKD A F+ A ++ A++ FN K
Sbjct: 621 DVLYLRQEDTSYLGPTSIQKLLKDTASRKFKDSAENHSIASEDLFNEK 764
>TC217543 similar to UP|Q940R0 (Q940R0) At1g29690/F15D2_24, partial (29%)
Length = 1093
Score = 201 bits (511), Expect = 8e-52
Identities = 121/287 (42%), Positives = 166/287 (57%), Gaps = 15/287 (5%)
Frame = +1
Query: 337 KPAIEELHQFLEFQLPRQWAPVFGELALGPDRKKSQSSSSLQFSFMGPKLYVNTSPVVVG 396
KP IE+L FL+FQ+ R WAP L ++K SLQFS MGPKL+V+ V VG
Sbjct: 124 KPPIEDLQYFLDFQITRVWAPEQNNL-----QRKEPVCQSLQFSLMGPKLFVSPDQVTVG 288
Query: 397 MKPVTGLRLYLEGKKSNCLAIHLQHLSSLPKTFQLKDETNRNV------SDASSERKYYE 450
KPVTGLRL LEG K N LAIHLQHL SLPK Q + + + + +++E
Sbjct: 289 RKPVTGLRLSLEGSKQNRLAIHLQHLVSLPKNLQPHWDAHMAIGAPKWHGPEEQDSRWFE 468
Query: 451 KVQWKSFSHICTAPVESYDDNA-------VVTGAHFEVGETGLKKVLFLRLHFCKVADAT 503
++WK+FSH+ TAP+E + + +VTGA V + G K VL L+L F KV T
Sbjct: 469 PIKWKNFSHVSTAPIEYTETSIGDLSGVHIVTGAQLGVWDFGAKNVLHLKLLFSKVPGCT 648
Query: 504 RVRAPEWDGSPG--LTQKSGMISTFISTRFSGPQKLPPPQPSDVNVNSALYPGGPPVPAQ 561
+R WD +P Q+S S+ ++ + S +K + S +++
Sbjct: 649 -IRRSVWDHNPSTPAAQRSDGASSSLTKKTSEDKK----EDSSIHIG------------- 774
Query: 562 APKLLKFVDTTEMTRGPQDLPGYWVVSGARLYVEKGKISLKVKYSLL 608
KL K VD TEM++GPQD+PG+W+V+GA+L VEKGKI L++KYSLL
Sbjct: 775 --KLAKIVDMTEMSKGPQDIPGHWLVTGAKLGVEKGKIVLRIKYSLL 909
>TC235110 similar to PIR|C86410|C86410 protein F3M18.18 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(17%)
Length = 754
Score = 174 bits (442), Expect = 8e-44
Identities = 90/167 (53%), Positives = 118/167 (69%), Gaps = 5/167 (2%)
Frame = +3
Query: 453 QWKSFSHICTAPVE----SYDDN-AVVTGAHFEVGETGLKKVLFLRLHFCKVADATRVRA 507
+W FSH+ TAPV+ S D++ A+VT A FEV G+KKVLFLRL F VA AT +R
Sbjct: 3 KWSMFSHVYTAPVQYSSSSMDESTAIVTKAWFEVKLVGMKKVLFLRLGFSTVASAT-IRR 179
Query: 508 PEWDGSPGLTQKSGMISTFISTRFSGPQKLPPPQPSDVNVNSALYPGGPPVPAQAPKLLK 567
EWDG ++KSG S +STR S + P +P+ V++NSA+Y GPPVP + PK+L
Sbjct: 180 SEWDGPSTTSRKSGFFSALMSTRLSKELQSPDQKPNKVDINSAIYNVGPPVPTRVPKMLS 359
Query: 568 FVDTTEMTRGPQDLPGYWVVSGARLYVEKGKISLKVKYSLLTVILQD 614
FVDT EM RGP+D PGYWVV+GA+L VE G+IS+K KYSLLT++ ++
Sbjct: 360 FVDTKEMVRGPEDTPGYWVVTGAKLCVEGGRISIKAKYSLLTILSEE 500
>TC220491
Length = 875
Score = 135 bits (341), Expect = 4e-32
Identities = 82/173 (47%), Positives = 104/173 (59%), Gaps = 10/173 (5%)
Frame = +2
Query: 337 KPAIEELHQFLEFQLPRQWAPVFGELALGPDRKKSQSSSSLQFSFMGPKLYVNTSPVVVG 396
KP ++L FLEFQ+PRQWAP+F +L L RKK SS SLQF FM PKL+V+ + VV
Sbjct: 359 KPPPDDLQCFLEFQIPRQWAPMFYDLPLRHQRKKC-SSPSLQFGFMFPKLHVSCAQVVSE 535
Query: 397 MKPVTGLRLYLEGKKSNCLAIHLQHLSSLPKTFQLKDETN--RNVSDASSERKYYEKVQW 454
KPV GLRLYLEG+KS+ LAIH+ HLSSLP T T+ R D S + E ++W
Sbjct: 536 QKPVVGLRLYLEGRKSDRLAIHVHHLSSLPNTMIYSSGTSSWRGSDDNESSDIFLEPIRW 715
Query: 455 KSFSHICTAPV--------ESYDDNAVVTGAHFEVGETGLKKVLFLRLHFCKV 499
K F+++CTA V E+ +VTGA T K VL LRL + +
Sbjct: 716 KGFANVCTAVVKHDPTWLQETSGGVYIVTGAQLISKGTWPKNVLHLRLLYTHI 874
>TC211049 similar to UP|Q9LQV3 (Q9LQV3) F10B6.18, partial (19%)
Length = 668
Score = 131 bits (330), Expect = 8e-31
Identities = 76/173 (43%), Positives = 109/173 (62%), Gaps = 11/173 (6%)
Frame = +1
Query: 447 KYYEKVQWKSFSHICTAPVESYDDN--------AVVTGAHFEVGETGLKKVLFLRLHFCK 498
+++E + K F H+CTAPV+ YD +VTGA V + + VL LRL F K
Sbjct: 7 RFFEAINGKKFYHVCTAPVK-YDPRWSSDKDVAFIVTGAQLHVKKHESRSVLHLRLLFSK 183
Query: 499 VADATRVRAPEWDGSPGLTQKSGMISTFISTRFSGP---QKLPPPQPSDVNVNSALYPGG 555
V++ V++ GS GL+Q+SG+ S IST SG QK P V ++S+++P G
Sbjct: 184 VSNCAVVKSSWTQGSSGLSQRSGIFSV-ISTSISGKDQNQKKPV-----VLLDSSVFPTG 345
Query: 556 PPVPAQAPKLLKFVDTTEMTRGPQDLPGYWVVSGARLYVEKGKISLKVKYSLL 608
PPVP Q KLLKF+DT+++ +GPQD PG+W+++GARL ++K KI L K+SLL
Sbjct: 346 PPVPVQTQKLLKFIDTSQLCKGPQDSPGHWLITGARLVLDKRKICLWAKFSLL 504
>TC209962
Length = 659
Score = 128 bits (322), Expect = 7e-30
Identities = 73/182 (40%), Positives = 109/182 (59%), Gaps = 10/182 (5%)
Frame = +3
Query: 437 RNVSDASSERKYYEKVQWKSFSHICTAPVESYDDN--------AVVTGAHFEVGETGLKK 488
R D S ++ E+++WK FS++CTA V+ +D N +VTGA + +
Sbjct: 24 RGSDDNESRDQFLERIRWKRFSNVCTAVVK-HDPNWLNNS*GVYIVTGAQLLSKGSWPRN 200
Query: 489 VLFLRLHFCKVADATRVRAPEWDGSPGLTQKSGMISTFISTRFSGPQ--KLPPPQPSDVN 546
VL LRL F + + +R EW +P ++KS + T +ST FS Q PP+ +
Sbjct: 201 VLHLRLLFAHTPNCS-IRKSEWTAAPEASRKSSFL-TNLSTTFSFTQHGNTGPPKQAPTV 374
Query: 547 VNSALYPGGPPVPAQAPKLLKFVDTTEMTRGPQDLPGYWVVSGARLYVEKGKISLKVKYS 606
+NS +YP GPPVP +A KLLK+V+T E+ RGP D PG+W+V+ A+L + GKI L+VK++
Sbjct: 375 LNSGVYPDGPPVPVRAGKLLKYVETAEVVRGPHDAPGHWLVTAAKLVTDGGKIGLQVKFA 554
Query: 607 LL 608
LL
Sbjct: 555 LL 560
>TC220973
Length = 986
Score = 113 bits (282), Expect = 3e-25
Identities = 69/209 (33%), Positives = 110/209 (52%), Gaps = 15/209 (7%)
Frame = +2
Query: 415 LAIHLQHLSSLPKTFQLKDETNRNVS------DASSERKYYEKVQWKSFSHICTAPVES- 467
L++H+QHL SLPK +++ + + +++E V+WK+FSH+ TAP+E+
Sbjct: 2 LSVHVQHLVSLPKILHPYWDSHVAIGAPKWQGPEEQDSRWFEPVKWKNFSHVSTAPIENP 181
Query: 468 ------YDDNAVVTGAHFEVGETGLKKVLFLRLHFCKVADATRVRAPEWDGSPGLTQKSG 521
+ +VTGA V + G + VL+++L + ++ T +R WD
Sbjct: 182 ETFIGDFSGVYIVTGAQLGVWDFGSRNVLYMKLLYSRLPGCT-IRRSLWD---------- 328
Query: 522 MISTFISTRFSGPQKLPPPQPSDVNVNSALYPGGPPVP--AQAPKLLKFVDTTEMTRGPQ 579
+P P VN + P + A A KL+K+VD ++MT+GPQ
Sbjct: 329 --------------HVPNKPPKTVNAENTSNPDNSTLRENATANKLVKYVDLSKMTKGPQ 466
Query: 580 DLPGYWVVSGARLYVEKGKISLKVKYSLL 608
D PG+W+V+G +L VEKGK+ L+VKYSLL
Sbjct: 467 DPPGHWLVTGGKLGVEKGKVVLRVKYSLL 553
>TC217541
Length = 746
Score = 110 bits (275), Expect = 2e-24
Identities = 65/173 (37%), Positives = 100/173 (57%), Gaps = 9/173 (5%)
Frame = +3
Query: 445 ERKYYEKVQWKSFSHICTAPVESYDDNA-------VVTGAHFEVGETGLKKVLFLRLHFC 497
+ +++E ++WK+FSH+ TAP+E + + +VTGA V + G K VL L+L F
Sbjct: 72 DSRWFEPIKWKNFSHVSTAPIEYTETSIGDLSGVHIVTGAQLGVWDFGAKNVLHLKLLFS 251
Query: 498 KVADATRVRAPEWDGSPG--LTQKSGMISTFISTRFSGPQKLPPPQPSDVNVNSALYPGG 555
KV T +R WD +P + Q+ S+ + + S +K + S +++
Sbjct: 252 KVPGCT-IRRSVWDHNPSAPVAQRPDGASSSLMKKTSEDKK----EDSSIHIG------- 395
Query: 556 PPVPAQAPKLLKFVDTTEMTRGPQDLPGYWVVSGARLYVEKGKISLKVKYSLL 608
KL K VD TEM++GPQD+PG+W+V+GA+L VEKGKI L++KYSLL
Sbjct: 396 --------KLAKIVDMTEMSKGPQDIPGHWLVTGAKLGVEKGKIVLRIKYSLL 530
>TC231359
Length = 683
Score = 106 bits (264), Expect = 3e-23
Identities = 49/104 (47%), Positives = 73/104 (70%)
Frame = +2
Query: 505 VRAPEWDGSPGLTQKSGMISTFISTRFSGPQKLPPPQPSDVNVNSALYPGGPPVPAQAPK 564
+R EW G+P ++KS + T +ST FS Q+ PP + + ++S +YP GPPVP ++ K
Sbjct: 26 IRKSEWGGAPEASRKSSFL-TNLSTTFSFTQQSPPQKQAPTVLDSGVYPDGPPVPVRSGK 202
Query: 565 LLKFVDTTEMTRGPQDLPGYWVVSGARLYVEKGKISLKVKYSLL 608
+LK+VDT+E RGP D PG+W+V+ A+L E GKI L+VK++LL
Sbjct: 203 MLKYVDTSEFVRGPHDAPGHWLVTAAKLVTEGGKIGLQVKFALL 334
>CA850828 weakly similar to GP|14209545|dbj P0481E12.26 {Oryza sativa
(japonica cultivar-group)}, partial (8%)
Length = 584
Score = 102 bits (253), Expect = 7e-22
Identities = 55/126 (43%), Positives = 80/126 (62%), Gaps = 2/126 (1%)
Frame = +2
Query: 485 GLKKVLFLRLHFCKVADATRVRAPEWDGSPGLTQKSGMISTFISTRFSGPQK--LPPPQP 542
G VL LRL F + + + +R EW +P ++KS + T +ST FS Q PP+
Sbjct: 59 GQGNVLHLRLLFTHIPNCS-IRKSEWTAAPEASRKSFL--TNLSTTFSFTQHGTTGPPKQ 229
Query: 543 SDVNVNSALYPGGPPVPAQAPKLLKFVDTTEMTRGPQDLPGYWVVSGARLYVEKGKISLK 602
+ +NS +YP GPPVP +A KLLK+V+T E+ RGP D PG+W+V+ A+L + GKI L+
Sbjct: 230 APTALNSGVYPDGPPVPVRAGKLLKYVETAEVVRGPHDAPGHWLVTAAKLVTDGGKIGLQ 409
Query: 603 VKYSLL 608
K++LL
Sbjct: 410 XKFALL 427
>CA936339
Length = 431
Score = 97.4 bits (241), Expect = 2e-20
Identities = 50/90 (55%), Positives = 60/90 (66%)
Frame = +1
Query: 337 KPAIEELHQFLEFQLPRQWAPVFGELALGPDRKKSQSSSSLQFSFMGPKLYVNTSPVVVG 396
KP IEEL FLEFQ+ WAP+ + ++K SLQFS MG KLYV+ + VG
Sbjct: 166 KPPIEELRYFLEFQIAHVWAPLHERIP--GQQRKEPICPSLQFSIMGQKLYVSQEQITVG 339
Query: 397 MKPVTGLRLYLEGKKSNCLAIHLQHLSSLP 426
PVTGLRL+LEG K N L++HLQHLSSLP
Sbjct: 340 RLPVTGLRLFLEGSKQNRLSVHLQHLSSLP 429
>BM953913
Length = 421
Score = 95.5 bits (236), Expect = 6e-20
Identities = 49/91 (53%), Positives = 61/91 (66%)
Frame = +2
Query: 337 KPAIEELHQFLEFQLPRQWAPVFGELALGPDRKKSQSSSSLQFSFMGPKLYVNTSPVVVG 396
KP IEEL FLEFQ+P WAP+ ++ ++K SLQFS MG KLYV+ + VG
Sbjct: 140 KPPIEELRYFLEFQIPCVWAPLQDKIP--GQQRKEPVCPSLQFSIMGQKLYVSQEQITVG 313
Query: 397 MKPVTGLRLYLEGKKSNCLAIHLQHLSSLPK 427
+PVTGL L LEG K N L++H+QHL SLPK
Sbjct: 314 RRPVTGLHLCLEGSKQNRLSVHVQHLVSLPK 406
>BF597072
Length = 316
Score = 90.5 bits (223), Expect = 2e-18
Identities = 44/102 (43%), Positives = 68/102 (66%)
Frame = +2
Query: 9 SVEDVIKSVGLGYDLTNDLRLKFCKYDSKLIAIDHDNLRTVELPGRVSIPNVPKSINCDK 68
+ + I S+GLG+D+T D+ L CK ++LI ++ + R +E+PG V+IPNVP SI C +
Sbjct: 11 AAHNAINSIGLGFDITLDITLDNCKKGARLIFVNEEQCRHLEIPGGVAIPNVPNSIKCVR 190
Query: 69 GDRMRLCSDVLSFQQMSEQFNQEVSLSGKIPTGHFNSAFQFS 110
G+ +RL SDVL+ QM E F+Q + LS + +GH ++F S
Sbjct: 191 GESIRLESDVLTQYQMMEHFDQ*MLLSRNLASGHLCASFGLS 316
>BM527909 weakly similar to PIR|C86410|C864 protein F3M18.18 [imported] -
Arabidopsis thaliana, partial (7%)
Length = 427
Score = 87.0 bits (214), Expect = 2e-17
Identities = 46/88 (52%), Positives = 59/88 (66%)
Frame = +1
Query: 76 SDVLSFQQMSEQFNQEVSLSGKIPTGHFNSAFQFSGVWQRDAANTKSLAFDGVSITLYNI 135
S+VL+ QQM E FNQE+ L G+ +GHF ++F S +D A+ KSLA+DG I Y +
Sbjct: 7 SEVLTLQQMLEHFNQEMCLGGQTASGHFCASFGLSCRNIKDLASIKSLAYDGWFIKRYAV 186
Query: 136 ALDKTHVVLSDHVKRAVPSSWDPAALAR 163
L++ L DHVK AVPSSWDP ALAR
Sbjct: 187 ELERYQGELLDHVKEAVPSSWDPEALAR 270
>TC230256
Length = 639
Score = 72.8 bits (177), Expect = 4e-13
Identities = 45/124 (36%), Positives = 64/124 (51%)
Frame = +1
Query: 485 GLKKVLFLRLHFCKVADATRVRAPEWDGSPGLTQKSGMISTFISTRFSGPQKLPPPQPSD 544
G + VL+++L + ++ T +R WD P PP+ +
Sbjct: 4 GSRNVLYMKLLYSRLPGCT-IRRSLWDHIPN----------------------KPPKTVN 114
Query: 545 VNVNSALYPGGPPVPAQAPKLLKFVDTTEMTRGPQDLPGYWVVSGARLYVEKGKISLKVK 604
S L A KL+K+VD +EMT+GPQD PG+W+V+G +L VEKGKI L+VK
Sbjct: 115 AGNTSNLDNSTLKENATGNKLVKYVDLSEMTKGPQDPPGHWLVTGGKLGVEKGKIVLRVK 294
Query: 605 YSLL 608
YSLL
Sbjct: 295 YSLL 306
>AW760549
Length = 296
Score = 57.4 bits (137), Expect(2) = 1e-11
Identities = 27/47 (57%), Positives = 35/47 (74%)
Frame = +3
Query: 337 KPAIEELHQFLEFQLPRQWAPVFGELALGPDRKKSQSSSSLQFSFMG 383
KP IE+LHQFLEFQLPR WAPV E++LG K Q ++ ++FS +G
Sbjct: 159 KPPIEDLHQFLEFQLPRHWAPVASEISLG-SHHKHQVNTWIRFSILG 296
Score = 30.8 bits (68), Expect(2) = 1e-11
Identities = 19/41 (46%), Positives = 25/41 (60%)
Frame = +1
Query: 291 MSGAKLFCLNLM*YQCHSYQLHLYLVE*MGVDI*LMP*ISI 331
++G L NLM*YQC H + +E +D+* MP*ISI
Sbjct: 28 VNGWILLTRNLM*YQCFFSL*HPFGIEAAEMDL*AMP*ISI 150
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,478,573
Number of Sequences: 63676
Number of extensions: 352672
Number of successful extensions: 2025
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 1955
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1999
length of query: 618
length of database: 12,639,632
effective HSP length: 103
effective length of query: 515
effective length of database: 6,081,004
effective search space: 3131717060
effective search space used: 3131717060
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148758.4


