

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.7 - phase: 0
(503 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
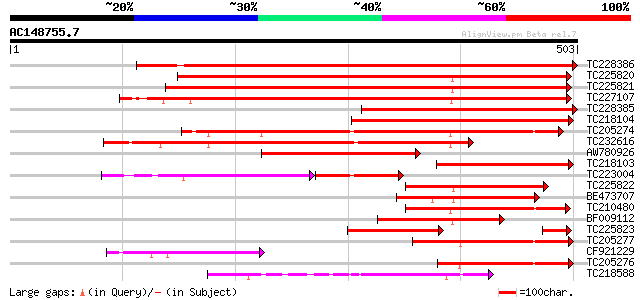
Score E
Sequences producing significant alignments: (bits) Value
TC228386 664 0.0
TC225820 weakly similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8,... 503 e-143
TC225821 weakly similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8,... 502 e-142
TC227107 similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partia... 472 e-133
TC228385 353 1e-97
TC218104 350 1e-96
TC205274 similar to GB|AAN73306.1|25141223|BT002309 At3g51670/T1... 330 9e-91
TC232616 similar to GB|AAD25756.1|4587525|T5I8 Contains the PF|0... 286 1e-77
AW780926 269 2e-72
TC218103 207 9e-54
TC223004 similar to GB|AAN73306.1|25141223|BT002309 At3g51670/T1... 133 2e-49
TC225822 homologue to PRF|1609232A.0|226866|1609232A 31kD glycop... 176 2e-44
BE473707 161 7e-40
TC210480 weakly similar to GB|AAD25756.1|4587525|T5I8 Contains t... 144 1e-34
BF009112 134 1e-31
TC225823 weakly similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8,... 130 2e-30
TC205277 similar to GB|AAN73306.1|25141223|BT002309 At3g51670/T1... 113 2e-25
CF921229 110 1e-24
TC205276 similar to GB|AAN73306.1|25141223|BT002309 At3g51670/T1... 95 6e-20
TC218588 UP|O48940 (O48940) Polyphosphoinositide binding protein... 85 7e-17
>TC228386
Length = 1517
Score = 664 bits (1713), Expect = 0.0
Identities = 328/391 (83%), Positives = 358/391 (90%)
Frame = +1
Query: 113 KKENVVSSVEDDGAKTVEAIEESIVAVSASVPPEQKPVVEKVEASLPLPPEQVSIYGIPL 172
K+ N ++V+DDGAKTVEAIEE++VAVS++V P+ + +AS PLPPE+VSI+GIPL
Sbjct: 1 KETNEEAAVDDDGAKTVEAIEETVVAVSSTVQPQAEE-----KASSPLPPEEVSIWGIPL 165
Query: 173 LADETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVYMHG 232
LADE SDVILLKFLRAR+F+VKEAFTM+KNTI WRKEFG+EELM+EKLGDELEKVV+MHG
Sbjct: 166 LADERSDVILLKFLRAREFRVKEAFTMLKNTIQWRKEFGMEELMEEKLGDELEKVVFMHG 345
Query: 233 FDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTI 292
FDKEGHPVCYNIY EFQNKELY KTFSDEEKR FL+WRIQFLEKSIR LDFN GG+CTI
Sbjct: 346 FDKEGHPVCYNIYEEFQNKELYKKTFSDEEKREKFLRWRIQFLEKSIRKLDFNPGGICTI 525
Query: 293 VHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLT 352
VHVNDLK+SPG KWELRQATK ALQL QDNYPEFVAKQVFINVPWWYLAVNRMISPFLT
Sbjct: 526 VHVNDLKNSPGLAKWELRQATKHALQLLQDNYPEFVAKQVFINVPWWYLAVNRMISPFLT 705
Query: 353 QRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDGEFGNSDSVTEITIRPASKHTV 412
QRTKSKFVFAGPSKSTETLL YIAPEQLPVKYGGLSKDGEFGN D+VTEIT+RPA+KH+V
Sbjct: 706 QRTKSKFVFAGPSKSTETLLRYIAPEQLPVKYGGLSKDGEFGNIDAVTEITVRPAAKHSV 885
Query: 413 EFPVTEKCLLSWEVRVIGWEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKIN 472
EF VTE CLLSWE+RVIGWEV YGAEFVPS+EGSYTVIVQKARKVASSEE VLCNSFK+
Sbjct: 886 EFSVTENCLLSWELRVIGWEVTYGAEFVPSSEGSYTVIVQKARKVASSEEPVLCNSFKVG 1065
Query: 473 EPGKVVLTIDNTSSRKKKLLYRLKTKTSLSD 503
EPGKVVLTIDNTSS+KKKLLYRLKTK S SD
Sbjct: 1066EPGKVVLTIDNTSSKKKKLLYRLKTKPSPSD 1158
>TC225820 weakly similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partial
(84%)
Length = 1376
Score = 503 bits (1296), Expect = e-143
Identities = 241/351 (68%), Positives = 291/351 (82%), Gaps = 2/351 (0%)
Frame = +2
Query: 150 VVEKVEASLPLPPEQVSIYGIPLLADETSDVILLKFLRARDFKVKEAFTMIKNTILWRKE 209
+ EK E+ +P+ PE+V I+GIPLL DE SDVILLKFLRARDFKVK+A +M++NT+ WRKE
Sbjct: 5 IEEKKESVVPISPEEVEIWGIPLLGDERSDVILLKFLRARDFKVKDALSMLRNTVRWRKE 184
Query: 210 FGIEELMDEKLGDELEKVVYMHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLK 269
FGIE L++E LG + +KVV+ HG DKEGHPV YN++GEF++KELYNKTF DEEKR+ ++
Sbjct: 185 FGIEGLVEEDLGSDWDKVVFSHGHDKEGHPVYYNVFGEFEDKELYNKTFWDEEKRNKLIR 364
Query: 270 WRIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVA 329
W IQ LEKS+R+LDF+ G+ TIV VNDLK+SPG GK ELRQAT Q LQLFQDNYPEFVA
Sbjct: 365 WMIQSLEKSVRSLDFSPTGISTIVQVNDLKNSPGLGKRELRQATNQVLQLFQDNYPEFVA 544
Query: 330 KQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSK 389
KQ+FINVPWWYLA +RMISPF TQRTKSKF+FAGPSKS TL YIAPE +PV+YGGLS+
Sbjct: 545 KQIFINVPWWYLAFSRMISPFFTQRTKSKFLFAGPSKSAHTLFQYIAPELVPVQYGGLSR 724
Query: 390 DG--EFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVRVIGWEVRYGAEFVPSNEGSY 447
+ EF SD VTE+TI+PA+KH VEFPV+EK WE+RV+GW+V YGAEFVP E Y
Sbjct: 725 EAEQEFTTSDPVTEVTIKPATKHAVEFPVSEKSHAVWEIRVVGWDVSYGAEFVPGAEDGY 904
Query: 448 TVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKLLYRLKTK 498
TVIVQK RK+ ++E V+ N+FKI EPGK+VLTIDN +S+KKKLLYR KTK
Sbjct: 905 TVIVQKNRKIGPADETVITNAFKIGEPGKIVLTIDNQTSKKKKLLYRSKTK 1057
>TC225821 weakly similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partial
(84%)
Length = 1480
Score = 502 bits (1292), Expect = e-142
Identities = 248/363 (68%), Positives = 297/363 (81%), Gaps = 3/363 (0%)
Frame = +2
Query: 139 VSASVPPEQKPV-VEKVEASLPLPPEQVSIYGIPLLADETSDVILLKFLRARDFKVKEAF 197
V V E+K V++ E + PE+V I+GIPLL DE SDVILLKFLRARDFKVKEA
Sbjct: 14 VEVEVREEKKESEVKEEEKGREVVPEEVEIWGIPLLGDERSDVILLKFLRARDFKVKEAL 193
Query: 198 TMIKNTILWRKEFGIEELMDEKLGDELEKVVYMHGFDKEGHPVCYNIYGEFQNKELYNKT 257
MI+NT+ WRKEFGIE L++E LG + EKVV+ G+DKEGHPV YN++GEF++KELY+KT
Sbjct: 194 NMIRNTVRWRKEFGIEGLVEEDLGSDWEKVVFKDGYDKEGHPVYYNVFGEFEDKELYSKT 373
Query: 258 FSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGPGKWELRQATKQAL 317
F DEEKR+ F++WRIQ LEKS+R+LDF+ G+ TIV VNDLK+SPG GK ELRQAT QAL
Sbjct: 374 FLDEEKRNKFIRWRIQSLEKSVRSLDFSPNGISTIVQVNDLKNSPGLGKRELRQATNQAL 553
Query: 318 QLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAP 377
QL QDNYPEFVAKQ+FINVPWWYLA +RMISPF TQRTKSKFVFAGPSKS +TL YIAP
Sbjct: 554 QLLQDNYPEFVAKQIFINVPWWYLAFSRMISPFFTQRTKSKFVFAGPSKSADTLFRYIAP 733
Query: 378 EQLPVKYGGLSKDG--EFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVRVIGWEVRY 435
E +PV+YGGLS++ EF ++ VTE TI+PA+KH+VEFPV+EK L WE+RV+GW+V Y
Sbjct: 734 ELVPVQYGGLSREAEQEFTSAYPVTEFTIKPATKHSVEFPVSEKSHLVWEIRVVGWDVSY 913
Query: 436 GAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKLLYRL 495
GAEFVPS E YTVIV K+RK+A ++E VL N FKI EPGK+VLTIDN +S+KKKLLYR
Sbjct: 914 GAEFVPSAEDGYTVIVHKSRKIAPADETVLTNGFKIGEPGKIVLTIDNQTSKKKKLLYRS 1093
Query: 496 KTK 498
KTK
Sbjct: 1094KTK 1102
>TC227107 similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partial (87%)
Length = 1785
Score = 472 bits (1215), Expect = e-133
Identities = 244/413 (59%), Positives = 306/413 (74%), Gaps = 12/413 (2%)
Frame = +1
Query: 98 VAAEPAQENKYQLEDKKENVVSSVEDDGAKTVEAIEES----IVAVSASVPPEQKPVVEK 153
VAAEP E+ Q DK+E EDD +K V + ES + S V +K +++
Sbjct: 91 VAAEPITEDLVQ--DKEE------EDDSSKIVIPVPESESLSLKEDSNRVSDSEKNAIDE 246
Query: 154 VEASLP--LPPEQVSIYGIPLLADETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFG 211
++ L L E+VSI+G+PL D+ +DVILLKFLRAR+ KVK+A M +NT+ WRK+F
Sbjct: 247 LKKLLKEELEDEEVSIWGVPLFKDDRTDVILLKFLRARELKVKDALVMFQNTLRWRKDFN 426
Query: 212 IEELMDEKLGDELEKVVYMHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWR 271
I+ L+DE LGD LEKVV+MHG +EGHPVCYN+YGEFQNK+LY+K FS ++ R+ FL+WR
Sbjct: 427 IDALLDEDLGDHLEKVVFMHGHGREGHPVCYNVYGEFQNKDLYHKAFSSQDNRNKFLRWR 606
Query: 272 IQFLEKSIRNLDFN-HGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAK 330
IQ LE+SIR+LDF G+ TI VNDLK+SPGP K ELR ATKQALQL QDNYPEFVAK
Sbjct: 607 IQLLERSIRHLDFTPSSGINTIFQVNDLKNSPGPAKRELRLATKQALQLLQDNYPEFVAK 786
Query: 331 QVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKD 390
QVFINVPWWYLA MI+PFLTQRTKSKFVFAGPSKS +TL YI+PEQ+PV+YGGLS D
Sbjct: 787 QVFINVPWWYLAFYTMINPFLTQRTKSKFVFAGPSKSPDTLFKYISPEQVPVQYGGLSVD 966
Query: 391 -----GEFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVRVIGWEVRYGAEFVPSNEG 445
+F SD VTEI I+P +K TVE + EKC++ WE+RV+GWEV Y AEF P E
Sbjct: 967 FCDCNPDFTMSDPVTEIPIKPTTKQTVEIAIYEKCIIVWELRVVGWEVSYNAEFKPDVED 1146
Query: 446 SYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKLLYRLKTK 498
+YTVI+QKA K++ ++E V+ NSFK+ E GK++LTIDN + +KK+LLYR K K
Sbjct: 1147AYTVIIQKATKMSPTDEPVVSNSFKVVELGKLLLTIDNPTLKKKRLLYRFKIK 1305
>TC228385
Length = 761
Score = 353 bits (905), Expect = 1e-97
Identities = 173/191 (90%), Positives = 181/191 (94%)
Frame = +2
Query: 313 TKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLL 372
TK ALQL QDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLL
Sbjct: 2 TKHALQLLQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLL 181
Query: 373 SYIAPEQLPVKYGGLSKDGEFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVRVIGWE 432
YIAPEQLPVKYGGL KDGEFGN+D+VTEIT+RPA+KHTVEF VTE CLLSWE+RVIGWE
Sbjct: 182 RYIAPEQLPVKYGGLGKDGEFGNTDAVTEITVRPAAKHTVEFSVTENCLLSWELRVIGWE 361
Query: 433 VRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKLL 492
V YGAEFVPS+EGSYTVIVQKARKVASSEE VLCNSFK+ EPGKVVLTIDNTSS+KKKLL
Sbjct: 362 VSYGAEFVPSSEGSYTVIVQKARKVASSEEPVLCNSFKVGEPGKVVLTIDNTSSKKKKLL 541
Query: 493 YRLKTKTSLSD 503
YRLKTK SLSD
Sbjct: 542 YRLKTKPSLSD 574
>TC218104
Length = 890
Score = 350 bits (897), Expect = 1e-96
Identities = 171/197 (86%), Positives = 182/197 (91%)
Frame = +3
Query: 304 PGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAG 363
P KWELRQATKQA QL QDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAG
Sbjct: 3 PAKWELRQATKQAXQLLQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAG 182
Query: 364 PSKSTETLLSYIAPEQLPVKYGGLSKDGEFGNSDSVTEITIRPASKHTVEFPVTEKCLLS 423
PSKS ETLL YIA EQLPVKYGGLSKDGEFG SD+VTEIT+RPA+KHTVEFPVTE LLS
Sbjct: 183 PSKSAETLLRYIAAEQLPVKYGGLSKDGEFGISDAVTEITVRPAAKHTVEFPVTENSLLS 362
Query: 424 WEVRVIGWEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDN 483
WE+RVIGW+V YGAEFVP++EGSYTVI+QKARKVASSEE VLCN++KI EPGKVVLTIDN
Sbjct: 363 WELRVIGWDVSYGAEFVPTSEGSYTVIIQKARKVASSEEPVLCNNYKIGEPGKVVLTIDN 542
Query: 484 TSSRKKKLLYRLKTKTS 500
SS+KKKLLYRLK K S
Sbjct: 543 QSSKKKKLLYRLKVKPS 593
>TC205274 similar to GB|AAN73306.1|25141223|BT002309 At3g51670/T18N14_50
{Arabidopsis thaliana;} , partial (79%)
Length = 1470
Score = 330 bits (846), Expect = 9e-91
Identities = 172/348 (49%), Positives = 239/348 (68%), Gaps = 9/348 (2%)
Frame = +3
Query: 153 KVEASLPLPPEQVSIYGIPLLAD---ETSDVILLKFLRARDFKVKEAFTMIKNTILWRKE 209
K+++S P S++G+PLL + + +DVILLKFLRARDF+V +A +M+ + WR E
Sbjct: 144 KLQSSYSSTP---SMWGVPLLNNNNADNADVILLKFLRARDFRVHDALSMLLKCLSWRTE 314
Query: 210 FGIEELMDEKLGD--ELEKVV-YMHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHN 266
FG + ++DE+LG ELE VV Y HG+D+EGHPVCYN YG F+++E+Y F DEEK
Sbjct: 315 FGADNIVDEELGGFKELEGVVAYTHGYDREGHPVCYNAYGVFKDREMYENVFGDEEKLKK 494
Query: 267 FLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPE 326
FL+WR+Q LE+ +R L F GGV +++ V DLKD P K ELR A+ Q L LFQDNYPE
Sbjct: 495 FLRWRVQVLERGVRMLHFKPGGVNSLIQVTDLKDMP---KRELRIASNQILSLFQDNYPE 665
Query: 327 FVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGG 386
VA+++FINVPW++ + M SPFLTQRTKSKFV + + ETL +I PE +PV+YGG
Sbjct: 666 MVARKIFINVPWYFSVLYSMFSPFLTQRTKSKFVISKEGNAAETLYKFIRPENIPVRYGG 845
Query: 387 LSK--DGEFGNSDSVTEITIRPASKHTVEFPVTEK-CLLSWEVRVIGWEVRYGAEFVPSN 443
LS+ D E G +E T++ ++ E ++W++ V GW++ Y AEFVP
Sbjct: 846 LSRPSDLENGPPKPASEFTVKGGEIVNIQIEGIESGATITWDIVVGGWDLEYSAEFVPIA 1025
Query: 444 EGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKL 491
+GSYT+ V KARK+ ++EEA+ NSF E GK+VL++DN++SRKKK+
Sbjct: 1026QGSYTLAVDKARKIEATEEAI-HNSFTSKEAGKMVLSVDNSASRKKKV 1166
>TC232616 similar to GB|AAD25756.1|4587525|T5I8 Contains the PF|00650
CRAL/TRIO phosphatidyl-inositol-transfer protein domain.
ESTs gb|T76582, gb|N06574 and gb|Z25700 come from this
gene. {Arabidopsis thaliana;} , partial (48%)
Length = 1214
Score = 286 bits (732), Expect = 1e-77
Identities = 156/342 (45%), Positives = 214/342 (61%), Gaps = 14/342 (4%)
Frame = +2
Query: 84 PPLIKEQKQSTTTTVAAEPAQENKYQLEDKKENVVSSVEDDGAKTVEAI--------EES 135
P + E K T ++ + N L D KE ++ + +K EAI ++
Sbjct: 200 PSSVDETKPKTVEKSSSYKEESN--YLSDLKEFERKALSELKSKLEEAILGNTLFDPKKE 373
Query: 136 IVAVSASVPPEQKPVVEKVEASLPLPPEQVSIYGIPLLAD---ETSDVILLKFLRARDFK 192
+ + E + E+ E + + VSI+G+ LL E +V+LLKFLRAR+FK
Sbjct: 374 ALPENEEKKNEGEEKEEEEEKRVEVEENDVSIWGVTLLPSKGAEGVEVVLLKFLRAREFK 553
Query: 193 VKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVYMHGFDKEGHPVCYNIYGEFQNKE 252
V +AF M+K T+ WRKE I+ ++DE G +L YM+G D EGHPVCYNI+G F+++E
Sbjct: 554 VNDAFEMLKKTLKWRKESKIDSVVDEDFGSDLASAAYMNGVDHEGHPVCYNIFGAFESEE 733
Query: 253 LYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGPGKWELRQA 312
Y KTF EEKR FL+WR Q +EK I+ L+ GGV +++ +NDLK+SPGP K LR A
Sbjct: 734 SYQKTFGTEEKRSEFLRWRCQLMEKGIQRLNLKPGGVSSLLQINDLKNSPGPSK--LRVA 907
Query: 313 TKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLL 372
TKQ L +FQDNYPE VAK +FINVP+WY A+N ++SPFLTQRTKSKFV A P+K TETL
Sbjct: 908 TKQTLAMFQDNYPEMVAKNIFINVPFWYYALNALLSPFLTQRTKSKFVVARPNKGTETLT 1087
Query: 373 SYIAPEQLPVKYGGLSK--DGEFGNSD-SVTEITIRPASKHT 411
YI E++PV YGG + D EF + D +V+E+ ++ S T
Sbjct: 1088KYIPIEEIPVHYGGSKRENDSEFSSQDVAVSELILKAGSTAT 1213
>AW780926
Length = 428
Score = 269 bits (687), Expect = 2e-72
Identities = 124/141 (87%), Positives = 131/141 (91%)
Frame = +2
Query: 224 LEKVVYMHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLD 283
LEK VYMHGFDKEGHPVCYNIYGEFQNKELY K+FSDEEKR+ FL+WRIQFLEKSIR LD
Sbjct: 5 LEKAVYMHGFDKEGHPVCYNIYGEFQNKELYKKSFSDEEKRYRFLRWRIQFLEKSIRKLD 184
Query: 284 FNHGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAV 343
FN GG+CTIV VNDL++SPGP KWELRQATKQALQL QDNYPEFVAKQVFINVPWWYLAV
Sbjct: 185 FNPGGICTIVQVNDLRNSPGPSKWELRQATKQALQLLQDNYPEFVAKQVFINVPWWYLAV 364
Query: 344 NRMISPFLTQRTKSKFVFAGP 364
NRMISPFLTQRTK + VFAGP
Sbjct: 365 NRMISPFLTQRTKXQXVFAGP 427
>TC218103
Length = 761
Score = 207 bits (527), Expect = 9e-54
Identities = 101/122 (82%), Positives = 112/122 (91%)
Frame = +2
Query: 379 QLPVKYGGLSKDGEFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVRVIGWEVRYGAE 438
QLPVKYGGLSKDGEFG SD+VTEIT+R A+KHTVEFPVTE LLSWE+RVIGW+V YGAE
Sbjct: 14 QLPVKYGGLSKDGEFGISDAVTEITVRSAAKHTVEFPVTENSLLSWELRVIGWDVSYGAE 193
Query: 439 FVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKLLYRLKTK 498
FVP++EGSYTVI+QKARKVASSEE VLCN++KI EPGKVVLTIDN SS+KKKLLYRLK K
Sbjct: 194 FVPTSEGSYTVIIQKARKVASSEEPVLCNNYKIGEPGKVVLTIDNQSSKKKKLLYRLKVK 373
Query: 499 TS 500
S
Sbjct: 374 PS 379
>TC223004 similar to GB|AAN73306.1|25141223|BT002309 At3g51670/T18N14_50
{Arabidopsis thaliana;} , partial (44%)
Length = 862
Score = 133 bits (334), Expect(2) = 2e-49
Identities = 83/196 (42%), Positives = 117/196 (59%), Gaps = 7/196 (3%)
Frame = +3
Query: 82 SAPPLIKEQKQSTTTTVAAEPAQENKYQLEDKKENVVSSVEDDGAKTVEAIEESIVAVSA 141
S+P ++ QK + A P + K+ +V+++ G +E VS
Sbjct: 84 SSPLSLQTQKTTFQELPEASP--------KPYKKGIVATLMGGGL-----FKEDNYFVSL 224
Query: 142 SVPPEQKPVVE---KVEASLPLPPEQVSIYGIPLLA-DETSDVILLKFLRARDFKVKEAF 197
E+K + E K++AS P S++GIPLL D+ +DVILLKFLRARDF+V +A
Sbjct: 225 LRSSEKKALQELKTKLKASFEDSPSDASMWGIPLLGGDDKADVILLKFLRARDFRVGDAH 404
Query: 198 TMIKNTILWRKEFGIEELMDEK-LG-DELEKVV-YMHGFDKEGHPVCYNIYGEFQNKELY 254
M+ + WRKEFG + +++E+ LG ELE VV YM G+DKEGHPVCYN YG F++KE+Y
Sbjct: 405 HMLMKCLSWRKEFGADTILEEEFLGLKELEGVVAYMQGYDKEGHPVCYNAYGVFKDKEMY 584
Query: 255 NKTFSDEEKRHNFLKW 270
+ F DEEK FL+W
Sbjct: 585 ERVFGDEEKLKKFLRW 632
Score = 81.3 bits (199), Expect(2) = 2e-49
Identities = 40/78 (51%), Positives = 53/78 (67%)
Frame = +1
Query: 272 IQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQ 331
+Q LE+ I+ L F GGV +++ V DLKD P K ELR A+ Q L LFQDNYPE VA++
Sbjct: 637 VQVLERGIKVLHFKPGGVNSLIQVTDLKDMP---KRELRVASNQILSLFQDNYPEMVARK 807
Query: 332 VFINVPWWYLAVNRMISP 349
+ INVPW++ + M SP
Sbjct: 808 IIINVPWYFSMLYSMFSP 861
>TC225822 homologue to PRF|1609232A.0|226866|1609232A 31kD glycoprotein.
{Glycine max;} , partial (44%)
Length = 820
Score = 176 bits (446), Expect = 2e-44
Identities = 84/129 (65%), Positives = 102/129 (78%), Gaps = 2/129 (1%)
Frame = -3
Query: 352 TQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDGE--FGNSDSVTEITIRPASK 409
TQRTKSKF+FAGPSKS TL YIAPE +PV+YGGLS++ E F SD VTE+TI+PA+K
Sbjct: 818 TQRTKSKFLFAGPSKSAHTLFQYIAPELVPVQYGGLSREAEQEFTTSDPVTEVTIKPATK 639
Query: 410 HTVEFPVTEKCLLSWEVRVIGWEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSF 469
H VEFPV+EK WE+RV+GW+V YGAEFVP E YTVIVQK RK+ ++E V+ N+F
Sbjct: 638 HAVEFPVSEKSHAVWEIRVVGWDVSYGAEFVPGAEDGYTVIVQKNRKIGPADETVITNAF 459
Query: 470 KINEPGKVV 478
KI EPGK+V
Sbjct: 458 KIGEPGKIV 432
>BE473707
Length = 470
Score = 161 bits (407), Expect = 7e-40
Identities = 84/155 (54%), Positives = 102/155 (65%), Gaps = 28/155 (18%)
Frame = +3
Query: 344 NRMISPFLTQRTKSKFVFAGPSKSTETLLS--------------------------YIAP 377
+RMISPF TQRTKSKF+FAGPSKS TL YIAP
Sbjct: 6 SRMISPFFTQRTKSKFLFAGPSKSAHTLFQ*LHFSFSSLLYMLLSIHYILLLSRYRYIAP 185
Query: 378 EQLPVKYGGLSKDGE--FGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVRVIGWEVRY 435
E +PV+YGGLS++ E F SD VTE+TI+PA+KH VEFPV+EK WE+RV+GW+V Y
Sbjct: 186 ELVPVQYGGLSREAEQEFTTSDPVTEVTIKPATKHAVEFPVSEKSHAVWEIRVVGWDVSY 365
Query: 436 GAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFK 470
GAEFVP E YTVIVQK RK+ ++E V+ N+FK
Sbjct: 366 GAEFVPGAEDGYTVIVQKNRKIGPADETVITNAFK 470
>TC210480 weakly similar to GB|AAD25756.1|4587525|T5I8 Contains the PF|00650
CRAL/TRIO phosphatidyl-inositol-transfer protein domain.
ESTs gb|T76582, gb|N06574 and gb|Z25700 come from this
gene. {Arabidopsis thaliana;} , partial (26%)
Length = 686
Score = 144 bits (362), Expect = 1e-34
Identities = 79/150 (52%), Positives = 104/150 (68%), Gaps = 4/150 (2%)
Frame = +3
Query: 352 TQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSK--DGEFGNSD-SVTEITIRPAS 408
TQRTKSKFV A +K ETL YI E++PV GG + D EF + D +V+E+ ++ S
Sbjct: 3 TQRTKSKFVVAXXNKVXETLTKYIPIEEIPVXXGGFKRENDXEFSSQDVAVSELILKAGS 182
Query: 409 KHTVEFPVTEKCL-LSWEVRVIGWEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCN 467
T+E P E L W++ V+GWE+ Y EFVP++EGSYTVIVQK +K+ S E V N
Sbjct: 183 TATIEIPALEVGYSLCWDLTVLGWELSYKEEFVPTDEGSYTVIVQKGKKMGSQEGPVR-N 359
Query: 468 SFKINEPGKVVLTIDNTSSRKKKLLYRLKT 497
+F+ NEPGKVVLTI NTS++KKK+LYR K+
Sbjct: 360 TFRNNEPGKVVLTIQNTSNKKKKVLYRYKS 449
>BF009112
Length = 346
Score = 134 bits (336), Expect = 1e-31
Identities = 63/115 (54%), Positives = 78/115 (67%), Gaps = 2/115 (1%)
Frame = +2
Query: 327 FVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGG 386
F +Q+FIN PWWYLA +RMI+PF TQRTKSKF FAGPSKS TL YI E P +YGG
Sbjct: 2 FGTRQIFINDPWWYLAFSRMITPFFTQRTKSKFFFAGPSKSAHTLFQYITTELAPAQYGG 181
Query: 387 LSKDG--EFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVRVIGWEVRYGAEF 439
LS++ E+ +D TE+T P +KH E PV EK WE+ ++ W+V GAEF
Sbjct: 182 LSREAYQEYTTADPSTEVTTTPTTKHAGEIPVYEKSHAFWEITMLDWDVSCGAEF 346
>TC225823 weakly similar to UP|Q94AG3 (Q94AG3) At1g72160/T9N14_8, partial
(27%)
Length = 728
Score = 130 bits (326), Expect = 2e-30
Identities = 63/86 (73%), Positives = 69/86 (79%)
Frame = +3
Query: 300 DSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKF 359
+S G K ELRQ T Q LQLFQDNY EFVAKQ+FINVPWWYLA +RMIS TQRTKSKF
Sbjct: 3 NSXGXXKRELRQXTNQVLQLFQDNYXEFVAKQIFINVPWWYLAFSRMISXXFTQRTKSKF 182
Query: 360 VFAGPSKSTETLLSYIAPEQLPVKYG 385
+FAGPSKS TL YIAPE +PV+YG
Sbjct: 183 LFAGPSKSAHTLFQYIAPELVPVQYG 260
Score = 46.6 bits (109), Expect = 3e-05
Identities = 21/26 (80%), Positives = 24/26 (91%)
Frame = +3
Query: 473 EPGKVVLTIDNTSSRKKKLLYRLKTK 498
EPGK+VLTIDN +S+KKKLLYR KTK
Sbjct: 261 EPGKIVLTIDNQTSKKKKLLYRSKTK 338
>TC205277 similar to GB|AAN73306.1|25141223|BT002309 At3g51670/T18N14_50
{Arabidopsis thaliana;} , partial (36%)
Length = 895
Score = 113 bits (283), Expect = 2e-25
Identities = 61/147 (41%), Positives = 92/147 (62%), Gaps = 4/147 (2%)
Frame = +2
Query: 358 KFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKDGEFGNSDS--VTEITIRPASKHTVEFP 415
KFV + + ETL ++ PE +PV+YGGL++ + N +E TI+ K ++
Sbjct: 2 KFVISKEGNAAETLYKFMRPEDIPVQYGGLNRPSDLQNGPPKPASEFTIKGGEKVNIQIE 181
Query: 416 VTEK-CLLSWEVRVIGWEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEP 474
E ++W++ V GW++ Y AEFVP+ EGSYT+ V+K RK+ +SEEA+ NSF E
Sbjct: 182 GIEAGATITWDIVVGGWDLEYSAEFVPNAEGSYTIAVEKPRKMGASEEAI-HNSFTSKES 358
Query: 475 GKVVLTIDNTSSRKKKL-LYRLKTKTS 500
GK+VL+ DNT+SR+KK+ YR + S
Sbjct: 359 GKMVLSADNTASRRKKVAAYRYVVRKS 439
>CF921229
Length = 680
Score = 110 bits (275), Expect = 1e-24
Identities = 71/154 (46%), Positives = 92/154 (59%), Gaps = 14/154 (9%)
Frame = -3
Query: 87 IKEQKQSTTTTVAAEPAQENKYQLEDKKENVVSSVEDD-----GAKTVEAIEESIVA--- 138
+ E+K+ T E +E + +E+KKE V+ + + G K VE IEE
Sbjct: 555 VTEEKKEVEVT---EEKKEAEVIVEEKKEVEVTEEKKEVEVTEGKKEVEVIEEKKETEVT 385
Query: 139 -----VSASVPPEQKPV-VEKVEASLPLPPEQVSIYGIPLLADETSDVILLKFLRARDFK 192
V V E+K V++ E + PE+V I+GIPLL DE SDVILLKFLRARDFK
Sbjct: 384 EEKKEVEVEVREEKKESEVKEEEKGQEVVPEEVEIWGIPLLGDERSDVILLKFLRARDFK 205
Query: 193 VKEAFTMIKNTILWRKEFGIEELMDEKLGDELEK 226
VKEA MI+NT+ WRKEFGIE L++E LG + EK
Sbjct: 204 VKEALNMIRNTVRWRKEFGIEGLVEEDLGSD*EK 103
>TC205276 similar to GB|AAN73306.1|25141223|BT002309 At3g51670/T18N14_50
{Arabidopsis thaliana;} , partial (31%)
Length = 685
Score = 95.1 bits (235), Expect = 6e-20
Identities = 52/125 (41%), Positives = 79/125 (62%), Gaps = 4/125 (3%)
Frame = +1
Query: 380 LPVKYGGLSKDGEFGNSD--SVTEITIRPASKHTVEFPVTEK-CLLSWEVRVIGWEVRYG 436
+PV YG L++ + N V+E I+ K ++ E ++W++ V GW++ Y
Sbjct: 1 IPVXYGXLNRPSDLQNXPPXXVSEFRIKGGEKVNIQIEGIEAGATITWDIVVGGWDLEYS 180
Query: 437 AEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKL-LYRL 495
AEFVP+ EGSYT+ V+K RK+ +SEEA+ NSF E GK+VL++DNT+SR+KK+ YR
Sbjct: 181 AEFVPNAEGSYTIAVEKPRKMGASEEAI-HNSFTSKESGKMVLSVDNTASRRKKVAAYRY 357
Query: 496 KTKTS 500
+ S
Sbjct: 358 VVRKS 372
>TC218588 UP|O48940 (O48940) Polyphosphoinositide binding protein Ssh2p,
complete
Length = 1030
Score = 85.1 bits (209), Expect = 7e-17
Identities = 75/276 (27%), Positives = 125/276 (45%), Gaps = 22/276 (7%)
Frame = +3
Query: 176 ETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEF------GIEELMDEKLGDELEKVVY 229
E D ++ +FLRARD V++A M+ + WR F + ++ +E D+ V+
Sbjct: 237 EEDDFMIRRFLRARDLDVEKASAMLLKYLKWRNSFVPNGSVSVSDVPNELAQDK----VF 404
Query: 230 MHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGV 289
M G DK G P+ F + NK DE KR + + L+K ++ G
Sbjct: 405 MQGHDKIGRPILM----VFGGRHFQNKDGLDEFKR-----FVVYVLDKVCASMP---PGQ 548
Query: 290 CTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISP 349
V + +LK G ++R AL + QD YPE + K +N P+ ++ V +++ P
Sbjct: 549 EKFVGIAELKGW-GYSNSDVR-GYLSALSILQDYYPERLGKLFIVNAPYIFMKVWQIVYP 722
Query: 350 FLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGG----------------LSKDGEF 393
F+ +TK K VF +K TLL + Q+P +GG L++ GE
Sbjct: 723 FIDNKTKKKIVFVEKNKVKSTLLEEMEESQVPEIFGGSLPLVPIQDS*LIK*PLTE*GEA 902
Query: 394 GNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVRVI 429
G ++ I+ S +P+T C +S++ V+
Sbjct: 903 GLKINIV-ISYLCVSMPFSFYPITLHCCISYQYSVV 1007
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.131 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,592,637
Number of Sequences: 63676
Number of extensions: 304809
Number of successful extensions: 1620
Number of sequences better than 10.0: 93
Number of HSP's better than 10.0 without gapping: 1548
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1591
length of query: 503
length of database: 12,639,632
effective HSP length: 101
effective length of query: 402
effective length of database: 6,208,356
effective search space: 2495759112
effective search space used: 2495759112
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148755.7


