

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.10 - phase: 0 /pseudo
(292 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
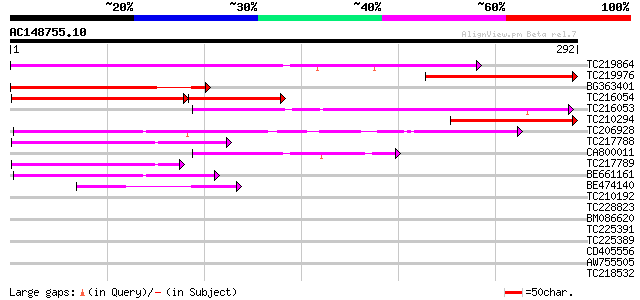
Score E
Sequences producing significant alignments: (bits) Value
TC219864 179 2e-45
TC219976 similar to UP|OAZ_HUMAN (P54368) Ornithine decarboxylas... 133 8e-32
BG363401 129 2e-30
TC216054 similar to UP|Q6NQK0 (Q6NQK0) At1g76970, partial (36%) 84 2e-28
TC216053 similar to UP|Q6NQK0 (Q6NQK0) At1g76970, partial (33%) 112 2e-25
TC210294 107 6e-24
TC206928 similar to UP|Q9LNC6 (Q9LNC6) F9P14.7 protein (At1g0621... 72 4e-13
TC217788 similar to UP|Q9LFL3 (Q9LFL3) TOM (Target of myb1)-like... 60 1e-09
CA800011 60 2e-09
TC217789 similar to UP|Q9LFL3 (Q9LFL3) TOM (Target of myb1)-like... 58 4e-09
BE661161 53 2e-07
BE474140 43 1e-04
TC210192 similar to UP|Q9SSY7 (Q9SSY7) Elicitor-responsive Dof p... 30 0.99
TC228823 similar to UP|Q9FL84 (Q9FL84) Arabidopsis thaliana geno... 30 0.99
BM086620 similar to SP|P83215|HSP3_O Sperm protamine P3 (Po3) (F... 29 2.2
TC225391 homologue to UP|Q84T14 (Q84T14) Vacuolar ATPase subunit... 29 2.2
TC225389 homologue to UP|Q84T14 (Q84T14) Vacuolar ATPase subunit... 29 2.9
CD405556 29 2.9
AW755505 29 2.9
TC218532 weakly similar to UP|Q9LP26 (Q9LP26) T9L6.9 protein, pa... 29 2.9
>TC219864
Length = 932
Score = 179 bits (453), Expect = 2e-45
Identities = 101/248 (40%), Positives = 146/248 (58%), Gaps = 5/248 (2%)
Frame = +3
Query: 1 MAAELVNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLGNKNPNAQLYAVM 60
M +V ATS+ L DW NIEI +++ D +AKDVVK IKKR+G+KN QL A+
Sbjct: 198 MVNSMVERATSDMLIGPDWAMNIEICDMLNHDPGQAKDVVKGIKKRIGSKNSKVQLLALT 377
Query: 61 LLEMLMNNIGDHINEQVVRAEVIPILVKIVKKKSDLPVREQIFLLLDATQTSLGGASGKF 120
LLE ++ N GD ++ V +V+ +VKIVKKK D V+E+I +L+D Q + GG ++
Sbjct: 378 LLETIIKNCGDIVHMHVAERDVLHEMVKIVKKKPDFHVKEKILVLVDTWQEAFGGPRARY 557
Query: 121 PQYYKAYYDLVSAGVQFPQRAQVVQSNRPSLQPNTTN---NVPKREPSPLRRGRVAQKAE 177
PQYY AY +L+ AG FPQR+ + + P P T + P+ P AQ +
Sbjct: 558 PQYYAAYQELLRAGAVFPQRS---EQSAPVFTPPQTQPLASYPQNIPDTNVDFYAAQSSA 728
Query: 178 SNTVPESRI--IQKASNVLEVLKEVLDAVDAKHPQGARDEFTLDLVEQCSFQKQRVMHLV 235
+ P + IQ A +++VL E+L+A+D + +G R E DLVEQC KQRV+HLV
Sbjct: 729 ESEFPTLNLTEIQNARGIMDVLAEMLNALDPSNKEGIRQEVIADLVEQCRTYKQRVVHLV 908
Query: 236 MASRDERI 243
++ DE +
Sbjct: 909 NSTSDESL 932
>TC219976 similar to UP|OAZ_HUMAN (P54368) Ornithine decarboxylase antizyme
(ODC-Az), partial (6%)
Length = 1046
Score = 133 bits (335), Expect = 8e-32
Identities = 67/78 (85%), Positives = 71/78 (90%)
Frame = +3
Query: 215 EFTLDLVEQCSFQKQRVMHLVMASRDERIVSRAIEVNEQLQKVLERHDDLLSSKDTTTVN 274
EFTLDLV QCSFQKQRVMHLVMASRDERIVSRAIE+NEQLQKVL RHDDLL+ + TTT +
Sbjct: 3 EFTLDLV*QCSFQKQRVMHLVMASRDERIVSRAIELNEQLQKVLARHDDLLAGRATTTAS 182
Query: 275 HFDHEEAEEEEEPEQLFR 292
FDHEEAEEEEEPEQL R
Sbjct: 183 RFDHEEAEEEEEPEQLVR 236
>BG363401
Length = 405
Score = 129 bits (324), Expect = 2e-30
Identities = 69/103 (66%), Positives = 79/103 (75%)
Frame = +3
Query: 1 MAAELVNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLGNKNPNAQLYAVM 60
MAAELVN ATSEKL+E DW KNIEI ELVA D+R+A+D VKAIKKRLG+K+PN QL+AVM
Sbjct: 147 MAAELVNGATSEKLAETDWTKNIEICELVAHDKRQARDAVKAIKKRLGSKHPNTQLFAVM 326
Query: 61 LLEMLMNNIGDHINEQVVRAEVIPILVKIVKKKSDLPVREQIF 103
LLEMLMNNIG+HI+EQ SDLPVRE+IF
Sbjct: 327 LLEMLMNNIGEHIHEQ-----------------SDLPVRERIF 404
>TC216054 similar to UP|Q6NQK0 (Q6NQK0) At1g76970, partial (36%)
Length = 713
Score = 84.0 bits (206), Expect(2) = 2e-28
Identities = 40/91 (43%), Positives = 58/91 (62%)
Frame = +1
Query: 2 AAELVNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLGNKNPNAQLYAVML 61
AA AT + LS DW NIE+ +++ D R+AKD +K +KKRL +KNP QL A+
Sbjct: 208 AAACAERATRDMLSGPDWAINIEVGDIINMDPRQAKDAIKILKKRLSSKNPQIQLLALFA 387
Query: 62 LEMLMNNIGDHINEQVVRAEVIPILVKIVKK 92
LE L N GD + +Q++ +++ +VKIVKK
Sbjct: 388 LETLSKNCGDSVFQQIIEQDILHEMVKIVKK 480
Score = 59.3 bits (142), Expect(2) = 2e-28
Identities = 27/50 (54%), Positives = 36/50 (72%)
Frame = +3
Query: 93 KSDLPVREQIFLLLDATQTSLGGASGKFPQYYKAYYDLVSAGVQFPQRAQ 142
+ DL VRE+I +L+D Q + GG SGK+PQY AY +L SAGV+FP R +
Sbjct: 474 EEDLRVREKILILIDTWQEAFGGPSGKYPQYLAAYNELKSAGVEFPPREE 623
>TC216053 similar to UP|Q6NQK0 (Q6NQK0) At1g76970, partial (33%)
Length = 1485
Score = 112 bits (281), Expect = 2e-25
Identities = 69/207 (33%), Positives = 115/207 (55%), Gaps = 11/207 (5%)
Frame = +3
Query: 95 DLPVREQIFLLLDATQTSLGGASGKFPQYYKAYYDLVSAGVQFPQRAQVVQSNRPSLQPN 154
DL VRE+I +L+D Q + GG +G +PQYY AY +L SAGV+FP R + ++ P P
Sbjct: 12 DLNVREKILILIDTWQEAFGGPTGVYPQYYAAYNELKSAGVEFPPRDE---NSVPFFTPA 182
Query: 155 TTNNVPKREPSPLRRGRVAQKAESNTVPESRI-IQKASNVLEVLKEVLDAVDAKHPQGAR 213
T + + + +S+ S + IQ A + +VL E+L A+ K +G +
Sbjct: 183 QTQPII-HSAAEYDDATIQASLQSDASDLSLLEIQNAQGLADVLMEMLSALSPKDREGVK 359
Query: 214 DEFTLDLVEQCSFQKQRVMHLVMASRDERIVSRAIEVNEQLQKVLERHDDLL-------- 265
+E +DLV+QC ++RVM LV + DE+++ + + +N+ LQ+VL RHDD++
Sbjct: 360 EEVIVDLVDQCRSYQKRVMLLVNNTTDEQLLGQGLALNDSLQRVLCRHDDIVKGTADSGA 539
Query: 266 --SSKDTTTVNHFDHEEAEEEEEPEQL 290
+ + + +HE+ E E++ QL
Sbjct: 540 REAETSVLPLVNVNHEDDESEDDFAQL 620
>TC210294
Length = 769
Score = 107 bits (267), Expect = 6e-24
Identities = 53/65 (81%), Positives = 57/65 (87%)
Frame = +3
Query: 228 KQRVMHLVMASRDERIVSRAIEVNEQLQKVLERHDDLLSSKDTTTVNHFDHEEAEEEEEP 287
KQRVMHLVMASRDERI+SRAIE+NEQLQKVL RHDDLL+ + T T FDHEEAEEEEEP
Sbjct: 3 KQRVMHLVMASRDERIISRAIELNEQLQKVLARHDDLLAGRVTMTTTRFDHEEAEEEEEP 182
Query: 288 EQLFR 292
EQL R
Sbjct: 183 EQLVR 197
>TC206928 similar to UP|Q9LNC6 (Q9LNC6) F9P14.7 protein (At1g06210/F9P14_4),
partial (58%)
Length = 1459
Score = 71.6 bits (174), Expect = 4e-13
Identities = 64/286 (22%), Positives = 131/286 (45%), Gaps = 24/286 (8%)
Frame = +1
Query: 3 AELVNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLGNKNPNAQLYAVMLL 62
+++V+ AT E + E +W N+ I ++ DQ +VVKAIK+++ +K+P Q ++ LL
Sbjct: 169 SKMVDEATLETMEEPNWGMNLRICGMINSDQFNGSEVVKAIKRKINHKSPVVQTLSLDLL 348
Query: 63 EMLMNNIGDHINEQVVRAEVIPILVKIV-----------------------KKKSDLPVR 99
E N D + ++ +V+ +++++ + + LPV
Sbjct: 349 EACAMNC-DKVFSEIASEKVLDEIIRLIDNPQAHHQTRSRAFQLIRAWGESEDLAYLPVF 525
Query: 100 EQIFLLLDATQTSLGGASGKFPQYYKAYYDLVSAGVQFPQRAQVVQSNRPSLQPNTTNNV 159
Q ++ L + A G P A V A P+R + Q+ +
Sbjct: 526 RQTYMSLKGRDEPVDMAGGNSPHVPYASESYVDA----PERYPIPQAELHDIDD------ 675
Query: 160 PKREPSPLRRGRVAQKAESNTVPESRIIQKASNVLEVLKEVLDAVDAKHPQGARDEFTLD 219
P S + V ++ E V A N LE+L +L++ DA+ P+ +++ T+
Sbjct: 676 PAAFSSNYQHISVEERKEHLVV--------ARNSLELLSSILNS-DAE-PKTLKEDLTVS 825
Query: 220 LVEQCSFQKQRVMHLV-MASRDERIVSRAIEVNEQLQKVLERHDDL 264
L+++C + +V + DE + A+ +N++LQ+++ ++++L
Sbjct: 826 LLDKCKQSLSIIKGIVESTTNDEATLFEALYLNDELQQIVSKYEEL 963
>TC217788 similar to UP|Q9LFL3 (Q9LFL3) TOM (Target of myb1)-like protein
(AT5g16880/F2K13_30), partial (52%)
Length = 1178
Score = 60.1 bits (144), Expect = 1e-09
Identities = 36/114 (31%), Positives = 66/114 (57%), Gaps = 1/114 (0%)
Frame = +1
Query: 2 AAELVNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLGNKNPNAQLYAVML 61
A +LV ATSE L E DW N+++ +L+ D+ + ++V+ IKKR+ K+P Q A++L
Sbjct: 328 ADKLVEDATSEALDEPDWALNLDLCDLINADKLSSVELVRGIKKRIVLKSPRVQYLALVL 507
Query: 62 LEMLMNNIGDHINEQVVRAEVIPILVKIV-KKKSDLPVREQIFLLLDATQTSLG 114
LE L+ N +E V V+ +V+++ ++ + R + ++++A S G
Sbjct: 508 LETLVKNCEKAFSE-VAAERVLDEMVRLIDDPQTVVNNRNKALMMIEAWAESTG 666
>CA800011
Length = 436
Score = 59.7 bits (143), Expect = 2e-09
Identities = 40/110 (36%), Positives = 58/110 (52%), Gaps = 3/110 (2%)
Frame = +3
Query: 95 DLPVREQIFLLLDATQTSLGGASGKFPQYYKAYYDLVSAGVQFPQRAQVVQSNRPSLQPN 154
DL VRE+I +L+D Q + GG SGK+PQY AY +L SAGV+FP R +++ P P
Sbjct: 123 DLRVREKILILIDTWQEAFGGPSGKYPQYLAAYNELKSAGVEFPPRE---ENSAPFFTPP 293
Query: 155 TTNNV---PKREPSPLRRGRVAQKAESNTVPESRIIQKASNVLEVLKEVL 201
T V + + A ++PE IQ A + +VL E++
Sbjct: 294 QTLPVHLAAAEYDDASIQASLHSDASGLSLPE---IQNAQGLADVLTEMV 434
>TC217789 similar to UP|Q9LFL3 (Q9LFL3) TOM (Target of myb1)-like protein
(AT5g16880/F2K13_30), partial (29%)
Length = 495
Score = 58.2 bits (139), Expect = 4e-09
Identities = 33/89 (37%), Positives = 53/89 (59%)
Frame = +3
Query: 2 AAELVNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLGNKNPNAQLYAVML 61
A +LV ATSE L E +W N+++ +LV D+ ++V+ IKKR+ K+P Q A++L
Sbjct: 165 ADKLVEDATSEALDEPEWALNLDLCDLVNTDKLNCVELVRGIKKRIILKSPRVQYLALVL 344
Query: 62 LEMLMNNIGDHINEQVVRAEVIPILVKIV 90
LE L+ N +E V V+ +VK++
Sbjct: 345 LETLVKNCEKAFSE-VAAERVLDEMVKLI 428
>BE661161
Length = 718
Score = 52.8 bits (125), Expect = 2e-07
Identities = 29/107 (27%), Positives = 60/107 (55%), Gaps = 1/107 (0%)
Frame = +2
Query: 3 AELVNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLGNKNPNAQLYAVMLL 62
+++V+ AT E + E +W N+ I ++ DQ +VVKAIK+++ +K+P Q ++ LL
Sbjct: 203 SKMVDEATLETMEEPNWGMNLRICSMINSDQFNGSEVVKAIKRKINHKSPVVQALSLDLL 382
Query: 63 EMLMNNIGDHINEQVVRAEVIPILVKIV-KKKSDLPVREQIFLLLDA 108
E N D + ++ +V+ +++++ ++ R + F L+ A
Sbjct: 383 EACAMNC-DKVFSEIASEKVLDEMIRLIDNPQAQHQTRSRAFQLIRA 520
>BE474140
Length = 427
Score = 43.1 bits (100), Expect = 1e-04
Identities = 28/85 (32%), Positives = 37/85 (42%)
Frame = +3
Query: 35 KAKDVVKAIKKRLGNKNPNAQLYAVMLLEMLMNNIGDHINEQVVRAEVIPILVKIVKKKS 94
+AKD +K +KKRL +KNP QL A+
Sbjct: 270 QAKDAIKILKKRLSSKNPQIQLLALF---------------------------------P 350
Query: 95 DLPVREQIFLLLDATQTSLGGASGK 119
DL VRE+I +L+D Q + GG SGK
Sbjct: 351 DLRVREKILILIDTWQEAFGGPSGK 425
Score = 27.7 bits (60), Expect = 6.4
Identities = 13/34 (38%), Positives = 19/34 (55%)
Frame = +1
Query: 2 AAELVNAATSEKLSEIDWMKNIEISELVARDQRK 35
AA ATS+ L DW NIE+ +++ D R+
Sbjct: 13 AAACAERATSDMLIGPDWAINIELCDIINMDPRE 114
>TC210192 similar to UP|Q9SSY7 (Q9SSY7) Elicitor-responsive Dof protein ERDP,
partial (24%)
Length = 1070
Score = 30.4 bits (67), Expect = 0.99
Identities = 20/80 (25%), Positives = 36/80 (45%), Gaps = 2/80 (2%)
Frame = -2
Query: 89 IVKKKSDLPVREQIFLLLDATQTSLGG--ASGKFPQYYKAYYDLVSAGVQFPQRAQVVQS 146
+V++ +L + +Q L++ + +G AS Y + + G Q + +V
Sbjct: 958 MVERSKELQLLQQESLIIVDAEVMMGNN*ASSSSSFNGIEYSNSFNGGSQVTEEDKVASD 779
Query: 147 NRPSLQPNTTNNVPKREPSP 166
RPS P+T NN+ SP
Sbjct: 778 QRPSSSPHTPNNLSNSSISP 719
>TC228823 similar to UP|Q9FL84 (Q9FL84) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K3K3, partial (25%)
Length = 675
Score = 30.4 bits (67), Expect = 0.99
Identities = 17/50 (34%), Positives = 25/50 (50%)
Frame = -1
Query: 243 IVSRAIEVNEQLQKVLERHDDLLSSKDTTTVNHFDHEEAEEEEEPEQLFR 292
IV R N K+ E+ + L + TTT H+ EE +P+QLF+
Sbjct: 462 IVHRKCSNNNNNNKLCEKLEALKNLIPTTTTTTTTHDGEEEAVKPDQLFK 313
>BM086620 similar to SP|P83215|HSP3_O Sperm protamine P3 (Po3) (Fragment).
[Octopus] {Octopus vulgaris}, partial (62%)
Length = 428
Score = 29.3 bits (64), Expect = 2.2
Identities = 16/41 (39%), Positives = 22/41 (53%), Gaps = 1/41 (2%)
Frame = +2
Query: 154 NTTNNVPKREPSPLRRGRVAQKAESNTVPES-RIIQKASNV 193
N T N PKR PSP R +V + N VP+ R +Q + +
Sbjct: 212 NLTTNTPKRRPSP--RTKVTRITRENLVPDKWREVQSETKI 328
>TC225391 homologue to UP|Q84T14 (Q84T14) Vacuolar ATPase subunit E
(Fragment), complete
Length = 1082
Score = 29.3 bits (64), Expect = 2.2
Identities = 21/113 (18%), Positives = 52/113 (45%), Gaps = 10/113 (8%)
Frame = +3
Query: 176 AESNTVPESRIIQKASNVLEVLKEVLDAVDAKHPQGARDEFTLDLVEQCSFQKQRVMHLV 235
A S + ++ + ++ +++ +++ + + A +EF ++ ++ K+++
Sbjct: 48 ASSPKMNDADVSKQIQQMVQFIRQEAEEKANEISVSAEEEFNIEKLQLVEADKKKIRQEY 227
Query: 236 MASRDERIVSRAIEVNEQLQ----KVLERHDDLLS------SKDTTTVNHFDH 278
+ + + IE + QL KVL+ DD++S SK+ V+H H
Sbjct: 228 ERKERQVEIRKKIEYSMQLNASRIKVLQAQDDVISSMKEAASKELLNVSHHRH 386
>TC225389 homologue to UP|Q84T14 (Q84T14) Vacuolar ATPase subunit E
(Fragment), complete
Length = 1222
Score = 28.9 bits (63), Expect = 2.9
Identities = 19/74 (25%), Positives = 37/74 (49%), Gaps = 10/74 (13%)
Frame = +1
Query: 212 ARDEFTLDLVEQCSFQKQRVMHLVMASRDERIVSRAIEVNEQLQ----KVLERHDDLLS- 266
A +EF ++ ++ +K+++ + + + IE + QL KVL+ DD++S
Sbjct: 193 AEEEFNIEKLQLVEAEKKKIRQEYERKERQVEIRKKIEYSMQLNASRIKVLQAQDDVISS 372
Query: 267 -----SKDTTTVNH 275
SK+ TV+H
Sbjct: 373 MKEAASKELLTVSH 414
>CD405556
Length = 611
Score = 28.9 bits (63), Expect = 2.9
Identities = 19/67 (28%), Positives = 36/67 (53%), Gaps = 6/67 (8%)
Frame = -2
Query: 131 VSAGVQFPQRAQVVQSNRPSLQPNTTNNVPKREPSPL-RRGRVAQ-----KAESNTVPES 184
VSAG P RA V ++P+ + ++ +R+PSP+ RGR+++ + +N S
Sbjct: 376 VSAGRSRP-RAVVTLPSKPNSEMQAPVSMSRRQPSPIANRGRLSEYTGKGRGHTNAADAS 200
Query: 185 RIIQKAS 191
++ + S
Sbjct: 199 EVVARRS 179
>AW755505
Length = 432
Score = 28.9 bits (63), Expect = 2.9
Identities = 21/79 (26%), Positives = 39/79 (48%), Gaps = 1/79 (1%)
Frame = +3
Query: 5 LVNAATSEKLSEIDWMKNIEISELVARDQRK-AKDVVKAIKKRLGNKNPNAQLYAVMLLE 63
L+++ATS++ K EI +L+ K++ + I KRL +KNP + A+ L++
Sbjct: 57 LIDSATSDEDKVTPVYKLEEICQLLLSSHATIVKELSEFILKRLDHKNPIVKHKALRLIK 236
Query: 64 MLMNNIGDHINEQVVRAEV 82
+ G ++ R V
Sbjct: 237 YAVGKCGVEFRREMQRHSV 293
>TC218532 weakly similar to UP|Q9LP26 (Q9LP26) T9L6.9 protein, partial (26%)
Length = 417
Score = 28.9 bits (63), Expect = 2.9
Identities = 29/111 (26%), Positives = 58/111 (52%), Gaps = 9/111 (8%)
Frame = +2
Query: 172 VAQKAESNTVPESRIIQKA-SNVLEVLKEVLDAVDAKHPQGARDEFTLDLVEQCSFQKQR 230
VA++A + ES I+++ ++V+E KEV DAV A + + DE L +QR
Sbjct: 20 VAREALPQAMEESSSIEESVASVVEEAKEVQDAVSAHISKASSDEEPL---------RQR 172
Query: 231 V------MHLVMASRDERIVSRAI--EVNEQLQKVLERHDDLLSSKDTTTV 273
V +H + +S D + ++ I + ++L + L+R ++ D++++
Sbjct: 173 VRRVDSRIHSLRSSLDSLVSTKQIPPSLADKLDEDLQRARCIIVDGDSSSL 325
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.130 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,212,428
Number of Sequences: 63676
Number of extensions: 97210
Number of successful extensions: 591
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 579
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 587
length of query: 292
length of database: 12,639,632
effective HSP length: 96
effective length of query: 196
effective length of database: 6,526,736
effective search space: 1279240256
effective search space used: 1279240256
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148755.10


