

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
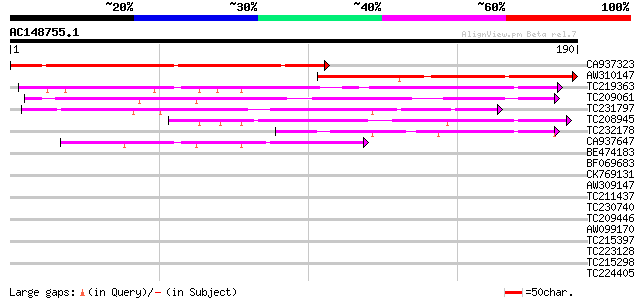
Query= AC148755.1 + phase: 0
(190 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CA937323 131 2e-31
AW310147 115 9e-27
TC219363 similar to UP|Q9FZA1 (Q9FZA1) F3H9.7 protein, partial (... 80 7e-16
TC209061 weakly similar to UP|Q9FNP0 (Q9FNP0) Gb|AAF27124.1, par... 75 1e-14
TC231797 similar to UP|Q9FZA1 (Q9FZA1) F3H9.7 protein, partial (... 58 2e-09
TC208945 similar to UP|Q9FHZ3 (Q9FHZ3) Gb|AAF34839.1 (AT5g53830/... 55 3e-08
TC232178 similar to UP|Q9FZA1 (Q9FZA1) F3H9.7 protein, partial (... 44 5e-05
CA937647 43 8e-05
BE474183 37 0.006
BF069683 homologue to GP|11385465|gb| glutathione S-transferase ... 37 0.006
CK769131 36 0.009
AW309147 36 0.009
TC211437 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%) 35 0.016
TC230740 similar to UP|Q9FZA1 (Q9FZA1) F3H9.7 protein, partial (... 35 0.021
TC209446 similar to UP|Q9LKA0 (Q9LKA0) Similarity to SF16 protei... 35 0.021
AW099170 similar to GP|14209533|dbj P0481E12.14 {Oryza sativa (j... 35 0.021
TC215397 weakly similar to UP|Q8S9J6 (Q8S9J6) AT5g10770/T30N20_4... 35 0.027
TC223128 similar to UP|WR27_ARATH (Q9FLX8) Probable WRKY transcr... 34 0.047
TC215298 similar to UP|Q40090 (Q40090) SPF1 protein, partial (21%) 33 0.079
TC224405 similar to UP|Q9LKA0 (Q9LKA0) Similarity to SF16 protei... 33 0.079
>CA937323
Length = 422
Score = 131 bits (330), Expect = 2e-31
Identities = 72/107 (67%), Positives = 80/107 (74%)
Frame = +2
Query: 1 MEKPPVHGSTTGDCKPLTTFVHTNTGAFREVVQRLTGPSEGNATKEEQTAKVAPITKRTP 60
MEKPP+HG+ + DCKPLTTFV TN+ AFREVVQRLTGPSE +A KE KVA + KRT
Sbjct: 110 MEKPPIHGAAS-DCKPLTTFVQTNSDAFREVVQRLTGPSEASAAKEGDAKKVATM-KRTT 283
Query: 61 PKLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSG 107
KLHERRK MKPK+EIVKP Y SP SKSR+SSFP SP SG
Sbjct: 284 SKLHERRKYMKPKVEIVKPTFNYKTGACLSP-SKSRSSSFPPSPGSG 421
>AW310147
Length = 441
Score = 115 bits (289), Expect = 9e-27
Identities = 66/88 (75%), Positives = 71/88 (80%), Gaps = 1/88 (1%)
Frame = -1
Query: 104 PVSGSGSSSLLPSPTTPSTLFSRLTI-MEDEKKQGSVINEINIEEEEKAIKERRFYLHPS 162
P GSGSSSLLPSPTTPSTLFS+L I +EDEKK+ SV EEEEKAIKERRFYLHPS
Sbjct: 441 PSPGSGSSSLLPSPTTPSTLFSQLKINLEDEKKEESV--NTAEEEEEKAIKERRFYLHPS 268
Query: 163 PRSKASGFNEPELLNLFPLASPKACEKV 190
PR+K G+ EPELLNLFPLAS A EKV
Sbjct: 267 PRAK-PGYTEPELLNLFPLASRNASEKV 187
>TC219363 similar to UP|Q9FZA1 (Q9FZA1) F3H9.7 protein, partial (44%)
Length = 1156
Score = 79.7 bits (195), Expect = 7e-16
Identities = 71/200 (35%), Positives = 97/200 (48%), Gaps = 18/200 (9%)
Frame = +1
Query: 4 PPVHGSTT--GDCKPL-TTFVHTNTGAFREVVQRLTGPSEGNATKEE------QTAKVAP 54
PP+ T D P TTFV +T F++VVQ LTG S+ + + + P
Sbjct: 310 PPLTPKTIPRSDSNPYPTTFVQADTSTFKQVVQMLTGSSDTTKQASQDPPPSSRNFNIPP 489
Query: 55 ITKRTPPK------LHERRK-CMKPKLEI--VKPNLQYHKQPGASPSSKSRNSSFPSSPV 105
I +TPPK L+ERR +K L + + PN ++ SPS RN SP
Sbjct: 490 I--KTPPKKQHGFKLYERRNNSLKNSLMLNTLMPNFAHNHNNNNSPSFSPRNMPEILSP- 660
Query: 106 SGSGSSSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRS 165
SLL P S S +T + D+ S + N EE+KAI E+ FYLHPSP +
Sbjct: 661 ------SLLDFP---SLALSPVTPLNDDPFDKSSPSLGNSSEEDKAIAEKGFYLHPSPMT 813
Query: 166 KASGFNEPELLNLFPLASPK 185
+EP+LL LFP+ SP+
Sbjct: 814 TPRD-SEPQLLPLFPVTSPR 870
>TC209061 weakly similar to UP|Q9FNP0 (Q9FNP0) Gb|AAF27124.1, partial (30%)
Length = 617
Score = 75.5 bits (184), Expect = 1e-14
Identities = 65/195 (33%), Positives = 84/195 (42%), Gaps = 16/195 (8%)
Frame = +3
Query: 6 VHGSTTGDCKPLTTFVHTNTGAFREVVQRLTGPSEGN--------------ATKEEQTAK 51
+H TT P TTFV N FR VVQ+LTG S+ A + + A
Sbjct: 24 LHSPTT----PNTTFVQANPSNFRAVVQKLTGASDDPSAHKLPLTLPTRLAAAQSHRPAS 191
Query: 52 VAPITKRTPP--KLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSG 109
+ P KLHERR +LE+ SP+S S S +
Sbjct: 192 AGEVIGPKKPNFKLHERRNAAAKRLELPIDTAMSMMMMSVSPTS--------SLVRSRTT 347
Query: 110 SSSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSKASG 169
+++L+ SP +P LF + EEEE+AI E+ FYLHPSPR+
Sbjct: 348 TTTLVASPVSPLELFLARASPRTPHE----------EEEERAIAEKGFYLHPSPRAS--- 488
Query: 170 FNEPELLNLFPLASP 184
PELL LFPL SP
Sbjct: 489 -QPPELLPLFPLHSP 530
>TC231797 similar to UP|Q9FZA1 (Q9FZA1) F3H9.7 protein, partial (37%)
Length = 658
Score = 58.2 bits (139), Expect = 2e-09
Identities = 58/170 (34%), Positives = 80/170 (46%), Gaps = 9/170 (5%)
Frame = +1
Query: 5 PVHGSTTGDCKPLTTFVHTNTGAFREVVQRLTGPSE----GNATKEEQT---AKVAPITK 57
PV S + + P TTFV +T +F++VVQ LTG ++ +A+ E + PI K
Sbjct: 139 PVTRSESANPYP-TTFVQADTTSFKQVVQMLTGSTQTAKQASASASEPAKPHTHIPPIKK 315
Query: 58 RTPPKLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSP 117
+T KL ERR + L I N SS S S +L+ SP
Sbjct: 316 QTGFKLFERRNSLSKNLSINPLN-------NVVVSSFSPRKHHEVLSPSILDFPALVLSP 474
Query: 118 TTP--STLFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRS 165
TP F+R + +E G V++ EE KAIKE+ F+LHPSP S
Sbjct: 475 VTPLIPDPFNRSRSL-NENGAGMVMDTAEATEE-KAIKEKGFFLHPSPAS 618
>TC208945 similar to UP|Q9FHZ3 (Q9FHZ3) Gb|AAF34839.1 (AT5g53830/MGN6_22),
partial (35%)
Length = 1127
Score = 54.7 bits (130), Expect = 3e-08
Identities = 53/147 (36%), Positives = 68/147 (46%), Gaps = 12/147 (8%)
Frame = +3
Query: 54 PITKRTPPK-----LHERRKC---MKPKLEI--VKPNLQYHKQPGASPSSKSRNSSFPSS 103
P K TP K L+ERR +K L I + PN H +S SS N S
Sbjct: 57 PPMKTTPKKQQGFKLYERRNNNNHLKNTLMINTLVPNFA-HSSGFSSSSSSPCNKPEILS 233
Query: 104 PVSGSGSSSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEINI--EEEEKAIKERRFYLHP 161
P S L SP TP + +D + S + + EEE+AI E+ FYLHP
Sbjct: 234 PSLLDFPSLTLSSPVTP--------LNDDPFDKSSSLPSLGSTSSEEERAIAEKGFYLHP 389
Query: 162 SPRSKASGFNEPELLNLFPLASPKACE 188
SP S +EP+LL LFP+ SP+ E
Sbjct: 390 SPMSTPRD-SEPQLLPLFPVTSPRVSE 467
>TC232178 similar to UP|Q9FZA1 (Q9FZA1) F3H9.7 protein, partial (27%)
Length = 575
Score = 43.9 bits (102), Expect = 5e-05
Identities = 39/99 (39%), Positives = 51/99 (51%), Gaps = 4/99 (4%)
Frame = +3
Query: 90 SPSSKSRNSSFPSSPVSGSGSSSLLPSPTTP--STLFSRLTIMEDEKKQGSVINE-INIE 146
SPSS+ PS +L+ SP TP F+R + + G+V+ + I
Sbjct: 60 SPSSRKPEVLSPSI----LDFPALVLSPVTPLIPDPFNRSRFLNEN---GTVLMDAIEAA 218
Query: 147 EEEKAIKERRFYLHPSPRSKASGFNEPELLNLFPL-ASP 184
EEKAIKE+ F+LHPSP S +P LL LFP ASP
Sbjct: 219 AEEKAIKEKGFFLHPSPASTPRD-EKPRLLPLFPTDASP 332
>CA937647
Length = 391
Score = 43.1 bits (100), Expect = 8e-05
Identities = 39/125 (31%), Positives = 54/125 (43%), Gaps = 22/125 (17%)
Frame = +3
Query: 18 TTFVHTNTGAFREVVQRLTG-------------PSEGNATKEEQTAKVAPITKRTPP--- 61
TTFV +T F++VVQ LTG P + ++ + + PI + PP
Sbjct: 18 TTFVQADTSTFKQVVQMLTGSSDTTKQASQDPQPPQPPSSSSSRNFNIPPI--KAPPKKQ 191
Query: 62 ----KLHERRKCMKPKLEI--VKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLP 115
KL+ERR +K L + + PN Y+ SPS RN+ S SL
Sbjct: 192 QGGFKLYERRNSLKNSLMLNTLMPNFAYNHN-NNSPSFSPRNNMPEILSPSLLDFPSLAL 368
Query: 116 SPTTP 120
SP TP
Sbjct: 369 SPVTP 383
>BE474183
Length = 357
Score = 37.0 bits (84), Expect = 0.006
Identities = 17/32 (53%), Positives = 23/32 (71%)
Frame = +2
Query: 154 ERRFYLHPSPRSKASGFNEPELLNLFPLASPK 185
E+ FYLHPSP + +EP+LL LFP+ SP+
Sbjct: 8 EKGFYLHPSPMTTPRD-SEPQLLPLFPVTSPR 100
>BF069683 homologue to GP|11385465|gb| glutathione S-transferase GST 25
{Glycine max}, partial (8%)
Length = 398
Score = 37.0 bits (84), Expect = 0.006
Identities = 26/73 (35%), Positives = 35/73 (47%), Gaps = 2/73 (2%)
Frame = +3
Query: 64 HERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSP--TTPS 121
H RR+ PK + PN + S S SS+ SS VS S S+SL+PSP +TPS
Sbjct: 39 HPRRRKKNPKRK-KSPNQRRRNYSARFSQSYSTRSSWSSSTVSSSPSTSLIPSPSASTPS 215
Query: 122 TLFSRLTIMEDEK 134
+ + K
Sbjct: 216 RTLGGALVPTESK 254
>CK769131
Length = 425
Score = 36.2 bits (82), Expect = 0.009
Identities = 31/93 (33%), Positives = 45/93 (48%), Gaps = 15/93 (16%)
Frame = +1
Query: 18 TTFVHTNTGAFREVVQRLTGPSE-------GNATKEEQT---AKVAPITKRTPPKLHERR 67
TTFV +T +F++VVQ LTG SE G ++K + K P K+ KL+ERR
Sbjct: 148 TTFVQADTSSFKQVVQMLTGSSETAKQAAAGGSSKPSNSIPPMKSIP-NKKHFSKLYERR 324
Query: 68 KCMKPKLEIVKPNLQYHK-----QPGASPSSKS 95
I P+L + + P +P+S S
Sbjct: 325 AYSLNLKNINPPHLVFSRTTTLNSPPKTPTSSS 423
>AW309147
Length = 332
Score = 36.2 bits (82), Expect = 0.009
Identities = 26/77 (33%), Positives = 35/77 (44%), Gaps = 6/77 (7%)
Frame = +3
Query: 91 PSSKSRNSSFPSSPVSGS------GSSSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEIN 144
PSS+ + P+SP + G S LLPS +P TL S T DE++ I
Sbjct: 87 PSSRRNPTPSPNSPPASYHFSHYFGDSLLLPSMASPDTLTSTPTSSMDERESLKNDTPIC 266
Query: 145 IEEEEKAIKERRFYLHP 161
+ KE+ FYL P
Sbjct: 267 SNACKDVTKEKTFYLQP 317
>TC211437 similar to UP|Q6VBJ4 (Q6VBJ4) Epa5p, partial (7%)
Length = 857
Score = 35.4 bits (80), Expect = 0.016
Identities = 19/38 (50%), Positives = 24/38 (63%)
Frame = -1
Query: 87 PGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTLF 124
P +SPSS +SS PSS SG SSS PS ++ +LF
Sbjct: 470 PPSSPSSPENSSSSPSSSSSGPSSSSSSPSSSSSVSLF 357
Score = 29.6 bits (65), Expect = 0.88
Identities = 17/38 (44%), Positives = 19/38 (49%)
Frame = -1
Query: 79 PNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPS 116
P+ P +SPSS S SS PS S SGS PS
Sbjct: 566 PSSSSSSSPLSSPSSSSSPSSLPSPLASSSGSPPSSPS 453
Score = 27.7 bits (60), Expect = 3.3
Identities = 18/37 (48%), Positives = 22/37 (58%), Gaps = 3/37 (8%)
Frame = -1
Query: 89 ASPSSKSRNSSFPSSPVSG---SGSSSLLPSPTTPST 122
+S SS S +SS SSP+S S S S LPSP S+
Sbjct: 587 SSSSSSSPSSSSSSSPLSSPSSSSSPSSLPSPLASSS 477
>TC230740 similar to UP|Q9FZA1 (Q9FZA1) F3H9.7 protein, partial (28%)
Length = 566
Score = 35.0 bits (79), Expect = 0.021
Identities = 42/153 (27%), Positives = 63/153 (40%), Gaps = 16/153 (10%)
Frame = +2
Query: 18 TTFVHTNTGAFREVVQRLTGPSE------------GNATKEEQTAKVAPITKRTPPKLHE 65
TTFV +T +F++VVQ LTG SE +++ + A P K P K +
Sbjct: 137 TTFVQADTTSFKQVVQMLTGSSETAKQAAAAAAAASSSSSSSKPANPIPPMKSIPNKKQQ 316
Query: 66 RR--KCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGS--GSSSLLPSPTTPS 121
K + + + H P S S NS + +S S +L+ SP TP
Sbjct: 317 PHFSKLYERRTNSLNLRNSLHINPLTSFFSNHTNSPRKADILSPSILDFPALVLSPVTP- 493
Query: 122 TLFSRLTIMEDEKKQGSVINEINIEEEEKAIKE 154
++ D + + I+ E E KAIKE
Sbjct: 494 -------LIPDPFDRSNA--AIDSEAEVKAIKE 565
>TC209446 similar to UP|Q9LKA0 (Q9LKA0) Similarity to SF16 protein, partial
(22%)
Length = 781
Score = 35.0 bits (79), Expect = 0.021
Identities = 22/61 (36%), Positives = 28/61 (45%), Gaps = 9/61 (14%)
Frame = +2
Query: 72 PKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVS---------GSGSSSLLPSPTTPST 122
P L I P HKQP S K +N+S P +PVS G+ + L PTT T
Sbjct: 257 PSLSIT-PKKSQHKQPSPSKVQKDKNTSTPKTPVSVKPPSANAKGTSKARRLSYPTTEKT 433
Query: 123 L 123
+
Sbjct: 434 V 436
>AW099170 similar to GP|14209533|dbj P0481E12.14 {Oryza sativa (japonica
cultivar-group)}, partial (10%)
Length = 108
Score = 35.0 bits (79), Expect = 0.021
Identities = 16/30 (53%), Positives = 20/30 (66%)
Frame = +3
Query: 157 FYLHPSPRSKASGFNEPELLNLFPLASPKA 186
F+ HPSPR K+ +P LL LFP SP+A
Sbjct: 3 FFFHPSPRDKS----QPRLLPLFPTTSPRA 80
>TC215397 weakly similar to UP|Q8S9J6 (Q8S9J6) AT5g10770/T30N20_40, partial
(76%)
Length = 1447
Score = 34.7 bits (78), Expect = 0.027
Identities = 23/93 (24%), Positives = 38/93 (40%)
Frame = +1
Query: 38 PSEGNATKEEQTAKVAPITKRTPPKLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRN 97
P + T A+ + PP P + PN P ASP +
Sbjct: 427 PPTSSTTSSSAAARTTRASSAAPPASLASAATPSPSSNKLPPNT-VKSSPTASPPPPAPP 603
Query: 98 SSFPSSPVSGSGSSSLLPSPTTPSTLFSRLTIM 130
++ PS+P + +SS PSP +P+ S ++I+
Sbjct: 604 ATSPSAPPPQAATSSTPPSPPSPAAPPSTVSIL 702
>TC223128 similar to UP|WR27_ARATH (Q9FLX8) Probable WRKY transcription
factor 27 (WRKY DNA-binding protein 27), partial (21%)
Length = 821
Score = 33.9 bits (76), Expect = 0.047
Identities = 19/40 (47%), Positives = 26/40 (64%)
Frame = -1
Query: 98 SSFPSSPVSGSGSSSLLPSPTTPSTLFSRLTIMEDEKKQG 137
SSFPS +SGSGS S + P+TL S + M+ +KK+G
Sbjct: 380 SSFPSVSISGSGSKSAVSVDLPPATLES-VVAMKKKKKKG 264
>TC215298 similar to UP|Q40090 (Q40090) SPF1 protein, partial (21%)
Length = 701
Score = 33.1 bits (74), Expect = 0.079
Identities = 35/135 (25%), Positives = 50/135 (36%), Gaps = 4/135 (2%)
Frame = +3
Query: 38 PSEGNATKEEQTAKVAPITKRTPPKLHERRKCMKPKLEI----VKPNLQYHKQPGASPSS 93
P G E+T P K TPP P L + + P+ + PG SP+
Sbjct: 159 PHGGGGGLSERTGSGVPKFKSTPP----------PSLPLSPPPISPSSYFSIPPGLSPAE 308
Query: 94 KSRNSSFPSSPVSGSGSSSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEINIEEEEKAIK 153
SPV SS++LPSPTT K S N+ ++EE+K
Sbjct: 309 ------LLDSPVL-LNSSNILPSPTT-----GAFVARSFNWKSSSGGNQRIVKEEDKGFS 452
Query: 154 ERRFYLHPSPRSKAS 168
F P + ++
Sbjct: 453 NFSFQTQQGPPASST 497
>TC224405 similar to UP|Q9LKA0 (Q9LKA0) Similarity to SF16 protein, partial
(14%)
Length = 764
Score = 33.1 bits (74), Expect = 0.079
Identities = 23/74 (31%), Positives = 30/74 (40%), Gaps = 18/74 (24%)
Frame = +1
Query: 72 PKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVS------------------GSGSSSL 113
P L + Q HKQP S K +N+S P +PVS G+ +
Sbjct: 175 PSLSVXXEKAQ-HKQPSPSKVQKDKNTSTPKTPVSFKPPSANAKGTPPSANAKGTSKARR 351
Query: 114 LPSPTTPSTLFSRL 127
L PTT TL R+
Sbjct: 352 LSYPTTEKTLVQRV 393
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.309 0.127 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,596,584
Number of Sequences: 63676
Number of extensions: 115381
Number of successful extensions: 1672
Number of sequences better than 10.0: 233
Number of HSP's better than 10.0 without gapping: 1496
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1621
length of query: 190
length of database: 12,639,632
effective HSP length: 92
effective length of query: 98
effective length of database: 6,781,440
effective search space: 664581120
effective search space used: 664581120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC148755.1


