

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148654.4 + phase: 0
(246 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
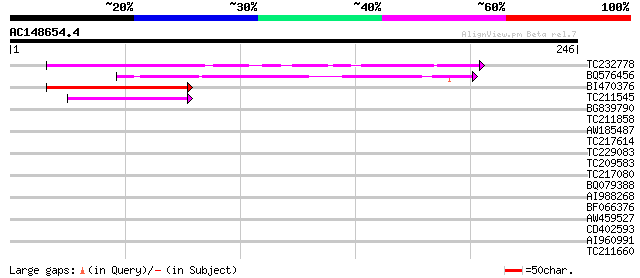
Score E
Sequences producing significant alignments: (bits) Value
TC232778 similar to UP|Q9LNG5 (Q9LNG5) F21D18.16, partial (5%) 78 4e-15
BQ576456 63 1e-10
BI470376 weakly similar to GP|14090338|dbj P0638D12.5 {Oryza sat... 59 3e-09
TC211545 weakly similar to UP|Q7XHQ1 (Q7XHQ1) Calcineurin-like p... 42 3e-04
BG839790 similar to PIR|D96521|D96 protein F21D18.16 [imported] ... 37 0.006
TC211858 35 0.024
AW185487 30 0.77
TC217614 similar to UP|Q7Y073 (Q7Y073) Latex cyanogenic beta glu... 30 1.3
TC229083 similar to UP|Q9LTZ9 (Q9LTZ9) Arabidopsis thaliana geno... 29 2.2
TC209583 29 2.2
TC217080 similar to UP|Q94EJ2 (Q94EJ2) At1g08460/T27G7_7 (HDA8),... 28 2.9
BQ079388 similar to PIR|S17744|S17 2'-hydroxyisoflavone reductas... 28 3.8
AI988268 GP|12620707|gb ID693 {Bradyrhizobium japonicum}, partia... 28 5.0
BF066376 28 5.0
AW459527 27 6.5
CD402593 27 6.5
AI960991 27 8.5
TC211660 similar to UP|Q711G4 (Q711G4) Cullin 3B (Fragment), par... 27 8.5
>TC232778 similar to UP|Q9LNG5 (Q9LNG5) F21D18.16, partial (5%)
Length = 746
Score = 77.8 bits (190), Expect = 4e-15
Identities = 58/190 (30%), Positives = 90/190 (46%)
Frame = -1
Query: 17 LRESGLHDLVYLGYATVPHALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEG 76
LR+ G + ++ +GY + AL+ E W PET TFH+ GE T+TL DV L L +G
Sbjct: 542 LRQCGFYWIMKMGYLKINAALISAFIERWRPETHTFHLRCGEATITLQDVLILLGLRTDG 363
Query: 77 RMLSHEKKMLKHKGATLLMRHLGVSQQEAEKICGQEYGGYISYPSLMDFYTTYLGRANLL 136
L + A L LGV QE G+ G + + + Y N+
Sbjct: 362 APLIGSTNL---DWADLCEELLGVRPQE-----GEIEGSVVK----LSWLAHYFSHINID 219
Query: 137 AGTEDPEELVRVRPYCVRCYLLYLVECLWFGDRSNKRIELVYLTTMEEGYAGMRNYSWGG 196
G + E+L R +R ++L + + F ++S+ R+ L YL + + + Y+WG
Sbjct: 218 EG--NVEQLQRF----IRAWILRFIGGVIFVNKSSSRVSLRYLQFLRD-FE*CSTYAWGP 60
Query: 197 MTLAYLYGEL 206
LAYLY E+
Sbjct: 59 AMLAYLYREM 30
>BQ576456
Length = 424
Score = 62.8 bits (151), Expect = 1e-10
Identities = 49/160 (30%), Positives = 74/160 (45%), Gaps = 3/160 (1%)
Frame = +1
Query: 47 PETSTFHMPLGEMTVTLDDVACLTHLPIEGRMLSHEKKMLKHKGATLLMRHLGVSQQEAE 106
P STF GE+T+TLDD+ACL HLPI G + E + + LLM L VS++EA
Sbjct: 7 PTPSTF*W--GELTITLDDMACLLHLPITGALHRFE-PLGVDEAVLLLMELLEVSREEAR 177
Query: 107 KICGQEYGGYISYPSLMDFYTTYLGRANLLAGTEDPEELVRVRPYCVRCYLLYLVECLWF 166
+ + Y+ L + Y + R VR YL +LV+C F
Sbjct: 178 AKIVRAHRAYVRLSWLREVYQSRC--------------QARCWIVAVRAYLFHLVDCTLF 315
Query: 167 GDRSNKRIELVYLTTMEEGYAGM---RNYSWGGMTLAYLY 203
++S + +V+L +G+ + Y WG L ++Y
Sbjct: 316 ANKSATHVHVVHL----KGF*DLC*SGGYGWGVAALVHMY 423
>BI470376 weakly similar to GP|14090338|dbj P0638D12.5 {Oryza sativa
(japonica cultivar-group)}, partial (5%)
Length = 429
Score = 58.5 bits (140), Expect = 3e-09
Identities = 28/63 (44%), Positives = 39/63 (61%)
Frame = -1
Query: 17 LRESGLHDLVYLGYATVPHALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEG 76
L+ SGL+ ++ L V AL+ E W PET TFH+ GE T+TL DV+ L +P++G
Sbjct: 393 LQISGLYPIMKLAQLKVNGALVNAFIERWRPETHTFHLKYGEATITLQDVSVLLGIPVDG 214
Query: 77 RML 79
R L
Sbjct: 213 RPL 205
>TC211545 weakly similar to UP|Q7XHQ1 (Q7XHQ1) Calcineurin-like
phosphoesterase-like, partial (11%)
Length = 1127
Score = 42.0 bits (97), Expect = 3e-04
Identities = 20/54 (37%), Positives = 31/54 (57%)
Frame = +3
Query: 26 VYLGYATVPHALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEGRML 79
+ +GY + +L+ L E P+T TFHM E T+TL DV+ L + ++G L
Sbjct: 3 IKMGYLKINASLITVLIER*RPKTHTFHMRCRECTITLQDVSVLLGMRVDGTPL 164
>BG839790 similar to PIR|D96521|D96 protein F21D18.16 [imported] -
Arabidopsis thaliana, partial (2%)
Length = 504
Score = 37.4 bits (85), Expect = 0.006
Identities = 15/38 (39%), Positives = 23/38 (60%)
Frame = -1
Query: 17 LRESGLHDLVYLGYATVPHALLMTLCEMWHPETSTFHM 54
LR+ G + ++ + Y + +L+ L E W PET TFHM
Sbjct: 258 LRQCGFY*IMKMSYLKINASLMTVLIERWRPETHTFHM 145
>TC211858
Length = 524
Score = 35.4 bits (80), Expect = 0.024
Identities = 20/54 (37%), Positives = 30/54 (55%), Gaps = 1/54 (1%)
Frame = -1
Query: 174 IELVYLTTMEE-GYAGMRNYSWGGMTLAYLYGELAEACRPGDRALGGSVTLLTV 226
+ +VYL + G +G Y+WG L ++Y +L EA R R + G +TLL V
Sbjct: 470 VHVVYLDAFRDLGQSG--GYAWGVAALVHMYNQLDEASRTTTRQIVGYLTLLQV 315
>AW185487
Length = 431
Score = 30.4 bits (67), Expect = 0.77
Identities = 12/27 (44%), Positives = 18/27 (66%)
Frame = +3
Query: 218 GGSVTLLTVRKLIFYFFVNFYFFCYIF 244
GGS+ +LT L+F+ F NFY ++F
Sbjct: 210 GGSIPVLTPTSLLFFLFTNFYPPLFLF 290
>TC217614 similar to UP|Q7Y073 (Q7Y073) Latex cyanogenic beta glucosidase,
partial (73%)
Length = 1382
Score = 29.6 bits (65), Expect = 1.3
Identities = 9/26 (34%), Positives = 17/26 (64%)
Frame = -1
Query: 157 LLYLVECLWFGDRSNKRIELVYLTTM 182
LLY + C+W+G N+ +L++ T +
Sbjct: 1154 LLYFIPCIWYGKTHNEESKLLFFTLL 1077
>TC229083 similar to UP|Q9LTZ9 (Q9LTZ9) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K15C23 (AT5g44670/K15C23_12),
partial (33%)
Length = 748
Score = 28.9 bits (63), Expect = 2.2
Identities = 13/26 (50%), Positives = 19/26 (73%)
Frame = +2
Query: 220 SVTLLTVRKLIFYFFVNFYFFCYIFR 245
+V L ++ LI +FF+NF F CYI+R
Sbjct: 647 AVLLFSLSCLIVFFFLNF-FMCYIYR 721
>TC209583
Length = 1349
Score = 28.9 bits (63), Expect = 2.2
Identities = 18/51 (35%), Positives = 28/51 (54%), Gaps = 2/51 (3%)
Frame = +2
Query: 124 DFYTTYLGRANLLAGTEDPEELVRVRPYCVRCY--LLYLVECLWFGDRSNK 172
D YT+ LG A LA ++DP CV+ + LLY++ WF + S++
Sbjct: 854 DDYTSVLGLAENLALSKDP---------CVKEFVKLLYMIMFRWFANESHR 979
>TC217080 similar to UP|Q94EJ2 (Q94EJ2) At1g08460/T27G7_7 (HDA8), partial
(88%)
Length = 1303
Score = 28.5 bits (62), Expect = 2.9
Identities = 11/19 (57%), Positives = 15/19 (78%)
Frame = -3
Query: 228 KLIFYFFVNFYFFCYIFRA 246
KL+ FF+NFY F Y+FR+
Sbjct: 347 KLLPPFFINFYQFIYVFRS 291
>BQ079388 similar to PIR|S17744|S17 2'-hydroxyisoflavone reductase (EC
1.3.1.45) - alfalfa, partial (34%)
Length = 421
Score = 28.1 bits (61), Expect = 3.8
Identities = 11/44 (25%), Positives = 22/44 (50%)
Frame = -3
Query: 43 EMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEGRMLSHEKKML 86
++W ++ T H+PL ++ T+ CL P++ H K +
Sbjct: 341 DLWQCQSHTLHLPLSAVSGTMPAGNCLEKKPLQ*TKKKHYKSRI 210
>AI988268 GP|12620707|gb ID693 {Bradyrhizobium japonicum}, partial (9%)
Length = 181
Score = 27.7 bits (60), Expect = 5.0
Identities = 8/20 (40%), Positives = 16/20 (80%)
Frame = -2
Query: 221 VTLLTVRKLIFYFFVNFYFF 240
+T+L + + IF+FF++F+ F
Sbjct: 150 ITILIILRFIFFFFISFFIF 91
>BF066376
Length = 407
Score = 27.7 bits (60), Expect = 5.0
Identities = 16/50 (32%), Positives = 22/50 (44%), Gaps = 10/50 (20%)
Frame = -2
Query: 42 CEMWHPETSTFHMPLGEMTVTLDD----------VACLTHLPIEGRMLSH 81
C W+ ETST P ++ L+ V+CL HLP+E H
Sbjct: 157 CLKWYLETSTHRGPF*LASLNLESRRDLISRRVRVSCLGHLPVEASPPMH 8
>AW459527
Length = 282
Score = 27.3 bits (59), Expect = 6.5
Identities = 10/38 (26%), Positives = 20/38 (52%)
Frame = -2
Query: 52 FHMPLGEMTVTLDDVACLTHLPIEGRMLSHEKKMLKHK 89
F P+ + D+ CL+H+P L H+ ++ +H+
Sbjct: 281 FKPPIHPPKYYMFDLTCLSHMPKSADKLEHQNRIKEHQ 168
>CD402593
Length = 639
Score = 27.3 bits (59), Expect = 6.5
Identities = 13/37 (35%), Positives = 21/37 (56%)
Frame = +2
Query: 208 EACRPGDRALGGSVTLLTVRKLIFYFFVNFYFFCYIF 244
EA +P +LG + L + +F+FF F+FF + F
Sbjct: 284 EAFQP---SLGQQIFLYDLSSCLFFFFFFFFFFFFFF 385
>AI960991
Length = 436
Score = 26.9 bits (58), Expect = 8.5
Identities = 9/14 (64%), Positives = 12/14 (85%)
Frame = -3
Query: 227 RKLIFYFFVNFYFF 240
RKL +FF+NF+FF
Sbjct: 107 RKLTLFFFINFFFF 66
>TC211660 similar to UP|Q711G4 (Q711G4) Cullin 3B (Fragment), partial (29%)
Length = 747
Score = 26.9 bits (58), Expect = 8.5
Identities = 10/16 (62%), Positives = 12/16 (74%)
Frame = -3
Query: 34 PHALLMTLCEMWHPET 49
PH L TLCE+ HP+T
Sbjct: 199 PHQPLSTLCEVCHPQT 152
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.326 0.142 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,297,990
Number of Sequences: 63676
Number of extensions: 205020
Number of successful extensions: 5069
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 2389
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4813
length of query: 246
length of database: 12,639,632
effective HSP length: 95
effective length of query: 151
effective length of database: 6,590,412
effective search space: 995152212
effective search space used: 995152212
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148654.4


