

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148651.4 + phase: 0 /pseudo
(764 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
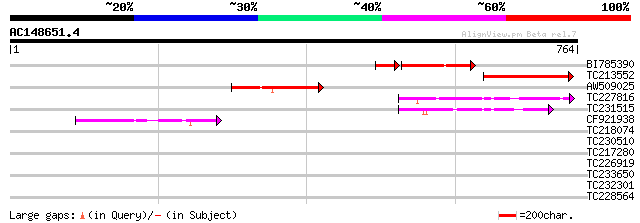
Score E
Sequences producing significant alignments: (bits) Value
BI785390 weakly similar to GP|13123673|gb molybdenum cofactor su... 157 2e-47
TC213552 similar to UP|Q9C5X8 (Q9C5X8) Molybdenum cofactor sulfu... 178 7e-45
AW509025 similar to GP|22128583|gb molybdenum cofactor sulfurase... 160 3e-39
TC227816 82 7e-16
TC231515 75 1e-13
CF921938 62 7e-10
TC218074 similar to UP|Q93WX6 (Q93WX6) NIFS-like protein CpNifsp... 40 0.005
TC230510 38 0.015
TC217280 31 1.8
TC226919 similar to UP|Q8LDZ7 (Q8LDZ7) Cinnamoyl CoA reductase-l... 31 2.3
TC233650 similar to UP|Q84JC9 (Q84JC9) BZIP transcription factor... 29 6.8
TC232301 weakly similar to UP|E2F4_HUMAN (Q16254) Transcription ... 29 6.8
TC228564 similar to UP|Q9XF14 (Q9XF14) Bundle sheath defective p... 29 6.8
>BI785390 weakly similar to GP|13123673|gb molybdenum cofactor sulfurase
{Arabidopsis thaliana}, partial (12%)
Length = 408
Score = 157 bits (398), Expect(2) = 2e-47
Identities = 77/100 (77%), Positives = 88/100 (88%)
Frame = +1
Query: 528 LKHDREWILKSLSGEILTLKRVPEMGLISSFIDLSQGMLFVESPHCKERLQIRLQLDFYD 587
L HDREWILKSL+GEILT K+VPEMG IS+FIDLSQGMLFVESP C+ERLQIRL+ D Y
Sbjct: 109 LTHDREWILKSLTGEILTQKKVPEMGFISTFIDLSQGMLFVESPRCEERLQIRLESDVY- 285
Query: 588 SAIQDIELQGQRYKVYSYDNETNSWFSKAIERPCTLLRYS 627
I++IEL GQRY+VYSYDNETNSWFS+AI + C+LLRYS
Sbjct: 286 GVIEEIELYGQRYEVYSYDNETNSWFSEAIGKTCSLLRYS 405
Score = 50.8 bits (120), Expect(2) = 2e-47
Identities = 28/33 (84%), Positives = 29/33 (87%)
Frame = +2
Query: 493 VSMILVIISNQLQCIR*NPVEASVQAVGLLATM 525
V ILVIISNQLQ I+*NPVE SVQAVGLLATM
Sbjct: 8 VLWILVIISNQLQYIQ*NPVEVSVQAVGLLATM 106
>TC213552 similar to UP|Q9C5X8 (Q9C5X8) Molybdenum cofactor sulfurase,
partial (12%)
Length = 704
Score = 178 bits (452), Expect = 7e-45
Identities = 87/121 (71%), Positives = 100/121 (81%)
Frame = +2
Query: 639 KDIVTCKDTNSAVSFANEGQFLLVSEESVSDLNKRLCSDVQMDMCETEIEINTNRFRPNL 698
K TC+D + ++FANE QFLLVSEESVSDLN+RL SDVQ + ++++ +RFRPNL
Sbjct: 5 KGAATCRDPKNKLNFANEAQFLLVSEESVSDLNRRLSSDVQKGIYGKVMQVSASRFRPNL 184
Query: 699 VVSGGRPYDEDGWSDIRIGNKYFRSLGGCNRCQVINLTLNAGQVQKSKEPLATLASYRRV 758
VVSGGRPY EDGW IRIG KYF SLGGCNRCQ+INLT+NAGQVQKS EPLATLASYRRV
Sbjct: 185 VVSGGRPYAEDGWRYIRIGIKYFSSLGGCNRCQIINLTINAGQVQKSNEPLATLASYRRV 364
Query: 759 K 759
K
Sbjct: 365 K 367
>AW509025 similar to GP|22128583|gb molybdenum cofactor sulfurase
{Lycopersicon esculentum}, partial (16%)
Length = 421
Score = 160 bits (404), Expect = 3e-39
Identities = 90/141 (63%), Positives = 98/141 (68%), Gaps = 16/141 (11%)
Frame = +1
Query: 299 KRREGIEELFEDGTVSFLSIASIRHGFKILNSLTVSAISRTYNISCPVYEENAF------ 352
KRREGIEELFED TVSFLSI SIRHGFKILNSLTVSAISR + S +Y
Sbjct: 1 KRREGIEELFEDVTVSFLSIVSIRHGFKILNSLTVSAISR-HIASLALYTRKMLLAMRHG 177
Query: 353 ----------GPKAWQWI*RLHPLWTP*FNGPDGSWYGYREVEKLASLSGIQLRTGCFCN 402
+ + + P+ + PDGSWYGYREVEKLASLSGIQLRTGCFCN
Sbjct: 178 NGSSVCILYGHHNSMKLCHEMGPIISFNLKRPDGSWYGYREVEKLASLSGIQLRTGCFCN 357
Query: 403 PGACAKYLGLSHMDLISNTEA 423
PGACAKYLGLSH+DLISNTEA
Sbjct: 358 PGACAKYLGLSHLDLISNTEA 420
>TC227816
Length = 1221
Score = 82.4 bits (202), Expect = 7e-16
Identities = 66/247 (26%), Positives = 107/247 (42%), Gaps = 10/247 (4%)
Frame = +2
Query: 524 TMGSLKHDREWILKSLSGEILTLK----------RVPEMGLISSFIDLSQGMLFVESPHC 573
T L+ DREW++ + G T + +P L+ F S + +++P
Sbjct: 254 TPAGLRWDREWVVVNSQGRACTQRVDPKLALVEVELPNDALVEDFEPTSDSYMVLKAPGM 433
Query: 574 KERLQIRLQLDFYDSAIQDIELQGQRYKVYSYDNETNSWFSKAIERPCTLLRYSGSSHDF 633
K + + L + + + E + WFS + +PC L+R++ +S
Sbjct: 434 KP---LNICLSKQHEVTDAVTVWEWTGSAWDEGAEASQWFSDFLGKPCQLVRFNSASEVR 604
Query: 634 VLDRTKDIVTCKDTNSAVSFANEGQFLLVSEESVSDLNKRLCSDVQMDMCETEIEINTNR 693
+D D V + F++ FLL+S+ES+ LN+ L + I NR
Sbjct: 605 QVD--PDYVRGQHRTY---FSDGYPFLLLSQESLDALNELL-----------KERIPINR 736
Query: 694 FRPNLVVSGGRPYDEDGWSDIRIGNKYFRSLGGCNRCQVINLTLNAGQVQKSKEPLATLA 753
FRPN++V G PY ED W++I+I F + C+RC+V + G EP TL
Sbjct: 737 FRPNILVEGCDPYSEDLWTEIKISRFSFLGVKLCSRCKVPTINQETGIA--GSEPTETLM 910
Query: 754 SYRRVKV 760
R KV
Sbjct: 911 KTRSGKV 931
>TC231515
Length = 720
Score = 75.1 bits (183), Expect = 1e-13
Identities = 58/220 (26%), Positives = 101/220 (45%), Gaps = 11/220 (5%)
Frame = +3
Query: 524 TMGSLKHDREWILKSLSGEILTLKRVPEMGLIS------SFID----LSQGMLFVESPHC 573
T + DR+W++ + G + T + P + L+ +F++ + V +P
Sbjct: 108 TPTGFRWDRQWMVVNSQGRMYTQRVEPRLALVEVELPSEAFLENWEPTQDSYMVVNAPGM 287
Query: 574 KE-RLQIRLQLDFYDSAIQDIELQGQRYKVYSYDNETNSWFSKAIERPCTLLRYSGSSHD 632
+ ++ + Q +A+ E G E + WFS + +PC L+R++ +S
Sbjct: 288 QPLKICLSQQGKEVANAVSVWEWTGS---ALDEGAEASQWFSDYLGKPCQLVRFNSASEV 458
Query: 633 FVLDRTKDIVTCKDTNSAVSFANEGQFLLVSEESVSDLNKRLCSDVQMDMCETEIEINTN 692
+D D V + + F + FLL S+ES+ +LN+ L V + N
Sbjct: 459 RPVD--PDYVKGQHQTT---FTDGYPFLLASQESLDELNEHLKEPVSI-----------N 590
Query: 693 RFRPNLVVSGGRPYDEDGWSDIRIGNKYFRSLGGCNRCQV 732
RFRPN++V G PY ED W+DI+I F + C RC++
Sbjct: 591 RFRPNILVEGCEPYSEDLWTDIKISKFLFSGVKLCYRCKI 710
>CF921938
Length = 905
Score = 62.4 bits (150), Expect = 7e-10
Identities = 54/204 (26%), Positives = 93/204 (45%), Gaps = 7/204 (3%)
Frame = -1
Query: 89 VLDYCNASPEDYKCIFTSGATAALKLVGEAFPWSCNSNFMYTME-NHNSVLGIREYALGQ 147
++ + N S DY +FT+ T+A KLV +++ + + + + +V + + +
Sbjct: 905 IMCFLNISENDYFMVFTANRTSAFKLVADSYQFQTSRRLLTVYDYESEAVEAMISSSKKR 726
Query: 148 GAAAIAVDIEDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEEPMECNFSGLRFDLDLAKI 207
GA AI+ + RI+ K K+ + +++K GL F L L+
Sbjct: 725 GARAISAEFSWPRLRIQTTKL-RKMIERKRKKKKRKGL--------------FVLPLSSR 591
Query: 208 IKEDSSKILGASVCKKGRWLVLIDAAKGSATMPPD-----LSKYPVDFVALSFYKLFG-Y 261
+ L S+ ++ W VL+DA A P D LS + DF+ SFYK+FG
Sbjct: 590 VTGARYPYLWMSIAQENGWHVLVDAC---ALGPKDMDCFGLSLFQPDFLICSFYKVFGEN 420
Query: 262 PTGLGALVVRNDAAKLLKKSYFSG 285
P+G G L ++ A L+ S +G
Sbjct: 419 PSGFGCLFIKKSAISSLESSSSAG 348
>TC218074 similar to UP|Q93WX6 (Q93WX6) NIFS-like protein CpNifsp precursor
(Cysteine desulfurase) (At1g08490/T27G7_14) , partial
(77%)
Length = 1302
Score = 39.7 bits (91), Expect = 0.005
Identities = 51/226 (22%), Positives = 100/226 (43%), Gaps = 7/226 (3%)
Frame = +2
Query: 74 SSAATHDIVRDARQQVLDYCNASPEDYKCIFTSGATAALKLVGEAFPWSCNSN------F 127
S+ AT++ AR++V + NA + + +FT A+ A+ LV A+ W ++
Sbjct: 14 SARATNEY-ESARRKVASFINAI-DSREIVFTKNASEAINLV--AYSWGLSNLKPDDEII 181
Query: 128 MYTMENHNSVLGIREYALGQGAAAIAVDI-EDVHPRIEGEKFPTKISLHQEQRRKVTGLQ 186
+ E+H++++ + A GA VD+ +D P I+ K+ ++ K+ +
Sbjct: 182 LTVAEHHSAIVPWQIVAQKTGAVLKFVDLNQDEIPDID------KLKEMLSRKTKIVVVH 343
Query: 187 EEEPMECNFSGLRFDLDLAKIIKEDSSKILGASVCKKGRWLVLIDAAKGSATMPPDLSKY 246
+ + +R D+A+ + +K VL+DA + M D+
Sbjct: 344 HVSNVLASVLPIR---DIAQWAHDVGAK-------------VLVDACQSVPHMMVDVQSL 475
Query: 247 PVDFVALSFYKLFGYPTGLGALVVRNDAAKLLKKSYFSGGTVAASI 292
DF+ S +K+ G PTG+G L + D + + GG + + +
Sbjct: 476 NADFLVASSHKMCG-PTGIGFLYGKIDLLSSM-PPFLGGGEMISDV 607
>TC230510
Length = 1198
Score = 38.1 bits (87), Expect = 0.015
Identities = 27/61 (44%), Positives = 34/61 (55%), Gaps = 3/61 (4%)
Frame = +1
Query: 228 VLIDA-AKGSATMPP-DLSKYPVDFVALSFYKLFG-YPTGLGALVVRNDAAKLLKKSYFS 284
VLIDA A G M LS + DF+ SFYK+FG P+G G L V+ A L+ S +
Sbjct: 4 VLIDACALGPKDMDSFGLSPFQPDFLICSFYKVFGENPSGFGCLFVKKSAISTLESSSCA 183
Query: 285 G 285
G
Sbjct: 184 G 186
>TC217280
Length = 1061
Score = 31.2 bits (69), Expect = 1.8
Identities = 12/25 (48%), Positives = 19/25 (76%)
Frame = +2
Query: 321 IRHGFKILNSLTVSAISRTYNISCP 345
IRHGF ++N L++S+ S ++ SCP
Sbjct: 23 IRHGFTVINQLSISSNSFLHSDSCP 97
>TC226919 similar to UP|Q8LDZ7 (Q8LDZ7) Cinnamoyl CoA reductase-like protein,
partial (52%)
Length = 1520
Score = 30.8 bits (68), Expect = 2.3
Identities = 30/117 (25%), Positives = 51/117 (42%), Gaps = 5/117 (4%)
Frame = +3
Query: 25 QIRATEFNRLQDLVYLDHAGATLYSELQMESVFKDLT----TNVYGNPHSQSDSSAATHD 80
++RATE N L+ A L +E F+ T+ + +P S + + +
Sbjct: 258 EVRATEGN-------LEVIMAKLTDVDGLEKAFQGCRGVFHTSAFTDPAGLSGYTKSMAE 416
Query: 81 IVRDARQQVLDYCNASPEDYKCIFTSGATAALKLVGEAFPWSCNSNFMYT-MENHNS 136
I A + V++ C +P +C+FTS +A + W NS +T + NH S
Sbjct: 417 IEVRAAENVMEACARTPSITRCVFTSSLSACV--------WQDNSQSDFTPVINHAS 563
>TC233650 similar to UP|Q84JC9 (Q84JC9) BZIP transcription factor, partial
(17%)
Length = 792
Score = 29.3 bits (64), Expect = 6.8
Identities = 16/52 (30%), Positives = 24/52 (45%), Gaps = 4/52 (7%)
Frame = -3
Query: 160 HPRIEGEKFPTKIS----LHQEQRRKVTGLQEEEPMECNFSGLRFDLDLAKI 207
HPR+ FP++ LH+ V GL EE+ CN F L+ + +
Sbjct: 592 HPRVMMGSFPSQNGDDEVLHRAAILGVGGLSEEDKQHCNSFNCLFSLEFSSV 437
>TC232301 weakly similar to UP|E2F4_HUMAN (Q16254) Transcription factor E2F4
(E2F-4), partial (5%)
Length = 587
Score = 29.3 bits (64), Expect = 6.8
Identities = 31/116 (26%), Positives = 44/116 (37%), Gaps = 18/116 (15%)
Frame = -1
Query: 644 CKDTNSAVSFANEGQFLLVSEESVSDLNKRLCSDVQM-DMC-------ETEIEINTNRFR 695
C D N E LLV S SD R CSD ++ C E E+ T F
Sbjct: 380 CSDLNR*AQ--QEAHLLLVPSRSCSDHRPRTCSDRRLFPTCLLFSS*LEDEVSSPTTSFP 207
Query: 696 PNLVVSGGRPYDEDGWS----------DIRIGNKYFRSLGGCNRCQVINLTLNAGQ 741
N + R D+D + +R + R+LG N+ + N +N G+
Sbjct: 206 SNFLFDSPRFDDDDDATLLLLESRVAFCVRGWRRKSRTLGEINKTRT*NRRVNRGR 39
>TC228564 similar to UP|Q9XF14 (Q9XF14) Bundle sheath defective protein 2,
partial (54%)
Length = 622
Score = 29.3 bits (64), Expect = 6.8
Identities = 17/44 (38%), Positives = 26/44 (58%), Gaps = 3/44 (6%)
Frame = +3
Query: 422 EAGHVCW---DDQDIISGKPIGAVRVSFGYMSTFEDAKKFIDFV 462
+AG +CW +DI+ G GA + G+MSTF+D K D++
Sbjct: 354 KAGGLCWLCRGKRDILCGSCNGAGFIG-GFMSTFDD*KNSPDWI 482
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.138 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 32,501,317
Number of Sequences: 63676
Number of extensions: 442373
Number of successful extensions: 1718
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 1703
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1713
length of query: 764
length of database: 12,639,632
effective HSP length: 105
effective length of query: 659
effective length of database: 5,953,652
effective search space: 3923456668
effective search space used: 3923456668
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC148651.4


