

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148651.11 - phase: 0
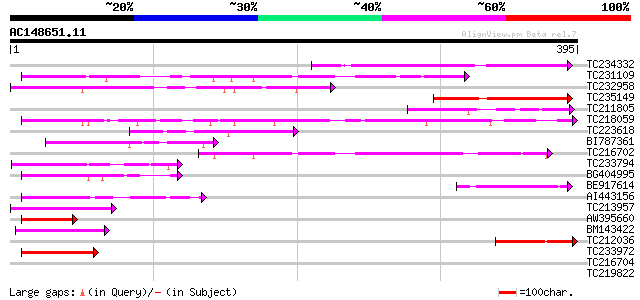
(395 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC234332 110 1e-24
TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, par... 102 2e-22
TC232958 79 5e-15
TC235149 74 9e-14
TC211805 similar to UP|Q9K6E7 (Q9K6E7) BH3782 protein, partial (... 74 1e-13
TC218059 weakly similar to UP|Q84KQ9 (Q84KQ9) F-box, partial (7%) 74 1e-13
TC223618 69 3e-12
BI787361 64 2e-10
TC216702 similar to UP|Q9LDP5 (Q9LDP5) FLORICAULA/LEAFY-like pro... 61 8e-10
TC233794 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {A... 57 2e-08
BG404995 similar to GP|22831068|dbj OJ1131_E05.32 {Oryza sativa ... 54 9e-08
BE917614 53 2e-07
AI443156 52 6e-07
TC213957 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {A... 49 5e-06
AW395660 48 9e-06
BM143422 47 1e-05
TC212036 43 2e-04
TC233972 weakly similar to GB|AAP37713.1|30725382|BT008354 At3g2... 42 6e-04
TC216704 similar to UP|Q7PIN1 (Q7PIN1) ENSANGP00000023661 (Fragm... 39 0.003
TC219822 homologue to UP|CENB_HUMAN (P07199) Major centromere au... 39 0.004
>TC234332
Length = 644
Score = 110 bits (274), Expect = 1e-24
Identities = 62/183 (33%), Positives = 96/183 (51%), Gaps = 1/183 (0%)
Frame = +2
Query: 211 TQNGVYFSSTVNWLALQDYFGLDYFHLNYSSITPEKYVILSLDLSTETYTQLLLPRGFNK 270
+Q+G TVNWLAL + DY + ++T + VI S DL E+Y LL+P G +
Sbjct: 11 SQDGASVRGTVNWLALPNSSS-DY---QWETVTIDDLVIFSYDLKNESYRYLLMPDGLLE 178
Query: 271 VSRHQPKLAVLMDCLCFGHDYEETYFVIWQMKDFGVQSSWIQLFKITYQNFFSYYCDFAR 330
V P+L VL CLC H + +F W MK+FGV+ SW + I+Y +
Sbjct: 179 VPHSPPELVVLKGCLCLSHRHGGNHFGFWLMKEFGVEKSWTRFLNISYDQLHIH------ 340
Query: 331 ESKWLDF-LPLCLSKNGDTLILANNQENEAFIYNRRDDRVEKIGITNKNMLWFEAWDYVE 389
+LD + LC+S++ ++L N + +YN+RD+ +E G +K F ++DY +
Sbjct: 341 -GGFLDHPVILCMSEDDGVVLLENGGHGKFILYNKRDNTIECYGELDKGRFQFLSYDYAQ 517
Query: 390 SLV 392
S V
Sbjct: 518 SFV 526
>TC231109 weakly similar to UP|Q9LUP7 (Q9LUP7) Gb|AAD25583.1, partial (8%)
Length = 1061
Score = 102 bits (255), Expect = 2e-22
Identities = 85/328 (25%), Positives = 157/328 (46%), Gaps = 16/328 (4%)
Frame = +3
Query: 9 LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQH-- 66
LP E++T+ILS LPVK ++R R K++ ++I HF+ HL S + +L++ R H
Sbjct: 93 LPVEVVTEILSRLPVKSVIRLRSTCKWWRSIIDSRHFVLFHLNKS--HSSLILRHRSHLY 266
Query: 67 --NFNSFDENVLNLPISLL-LENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLCLD 123
+ S ++N + L L+ NS+ + GS NGL+C+
Sbjct: 267 SLDLKSPEQNPVELSHPLMCYSNSIKVL--------------------GSSNGLLCI--- 377
Query: 124 IDTSHGSRLCLWNPATR-----TKSEFDLASQECF---VFAFGYDNLNGNYKV--IAFDI 173
++ + LWNP R F F V+ FG+ + + +YK+ I + +
Sbjct: 378 --SNVADDIALWNPFLRKHRILPADRFHRPQSSLFAARVYGFGHHSPSNDYKLLSITYFV 551
Query: 174 KVKSGNARSVVKVFSMRDNCWRNIQCFPVLPLYMFVSTQNGVYFSSTVNWLALQDYFGLD 233
++ S V++++++ + W+N+ P +P + + GV+ S +++WL +
Sbjct: 552 DLQKRTFDSQVQLYTLKSDSWKNL---PSMPYALCCARTMGVFVSGSLHWLVTR------ 704
Query: 234 YFHLNYSSITP-EKYVILSLDLSTETYTQLLLPRGFNKVSRHQPKLAVLMDCLCFGHDYE 292
+ P E +I+S DL+ ET+ ++ LP N ++A+L CLC ++
Sbjct: 705 -------KLQPHEPDLIVSFDLTRETFHEVPLPVTVN--GDFDMQVALLGGCLCV-VEHR 854
Query: 293 ETYFVIWQMKDFGVQSSWIQLFKITYQN 320
T F +W M+ +G ++SW +LF + N
Sbjct: 855 GTGFDVWVMRVYGSRNSWEKLFTLLENN 938
>TC232958
Length = 758
Score = 78.6 bits (192), Expect = 5e-15
Identities = 70/241 (29%), Positives = 107/241 (44%), Gaps = 14/241 (5%)
Frame = +1
Query: 1 MEDRHQSVLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMH--LKNSARNPN 58
+E R + VLP +L+T+IL LPVK L+RF+ V K + LISDP F + H L + +
Sbjct: 37 VERRKKMVLPQDLITEILLRLPVKSLVRFKSVCKSWLFLISDPRFAKSHFDLFAALADRI 216
Query: 59 LMVIARQHNFNSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLI 118
L + + S D N S + ++ PY+ GSC G I
Sbjct: 217 LFIASSAPELRSIDFNASLHDDSASVAVTVDLPAPKPYFHFVE--------IIGSCRGFI 372
Query: 119 CL-CLDIDTSHGSRLCLWNPATRTKSEFDLA----SQECFVF----AFGYDNLNGNYKVI 169
L CL S LC+WNP T L+ +++ F FGYD ++ V+
Sbjct: 373 LLHCL-------SHLCVWNPTTGVHKVVPLSPIFFNKDAVFFTLLCGFGYDPSTDDFLVV 531
Query: 170 AFDIKVKSGNARSVVKVFSMRDNCWRNIQ--CFPVLPL-YMFVSTQNGVYFSSTVNWLAL 226
K + + ++FS+R N W+ I+ FP Y Q G + + ++WLA
Sbjct: 532 HACYNPK--HQANCAEIFSLRANAWKGIEGIHFPYTHFRYTNRYNQFGSFLNGAIHWLAF 705
Query: 227 Q 227
+
Sbjct: 706 R 708
>TC235149
Length = 435
Score = 74.3 bits (181), Expect = 9e-14
Identities = 37/98 (37%), Positives = 62/98 (62%), Gaps = 1/98 (1%)
Frame = +2
Query: 296 FVIWQMKDFGVQSSWIQLFKITYQNFFSYYCDFARESKWLDFL-PLCLSKNGDTLILANN 354
FV+W ++FGV+ SW +L ++Y++F ++ C + F+ PLC+S+N D L+LAN+
Sbjct: 2 FVVWLTREFGVERSWTRLLNVSYEHFRNHGCP-----PYYRFVTPLCMSENEDVLLLAND 166
Query: 355 QENEAFIYNRRDDRVEKIGITNKNMLWFEAWDYVESLV 392
+ +E YN RD+R+++I + F + DYV SLV
Sbjct: 167 EGSEFVFYNLRDNRIDRIQDFDSYKFSFLSHDYVPSLV 280
>TC211805 similar to UP|Q9K6E7 (Q9K6E7) BH3782 protein, partial (13%)
Length = 693
Score = 73.9 bits (180), Expect = 1e-13
Identities = 44/119 (36%), Positives = 63/119 (51%), Gaps = 3/119 (2%)
Frame = +1
Query: 278 LAVLMDCLCFGHDYEETYFVIWQMKDFGVQSSWIQLFKITY---QNFFSYYCDFARESKW 334
L VL CLC +DY++T+FV+W MKD+G + SW++L I Y FSY
Sbjct: 4 LGVLQGCLCMNYDYKKTHFVVWMMKDYGARESWVKLVSIPYVPNPENFSYSG-------- 159
Query: 335 LDFLPLCLSKNGDTLILANNQENEAFIYNRRDDRVEKIGITNKNMLWFEAWDYVESLVS 393
P +S+NG+ L++ E + +YN RD+ K WF+A YVE+LVS
Sbjct: 160 ----PYYISENGEVLLMF---EFDLILYNPRDNSF-KYPKIESGKGWFDAEVYVETLVS 312
>TC218059 weakly similar to UP|Q84KQ9 (Q84KQ9) F-box, partial (7%)
Length = 1185
Score = 73.9 bits (180), Expect = 1e-13
Identities = 99/421 (23%), Positives = 178/421 (41%), Gaps = 34/421 (8%)
Frame = +3
Query: 9 LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMH--LKNS--ARNPNLMVIAR 64
LP EL++ +LS LP K L+ +CV K + LI+DPHF+ + + NS ++ +L+VI R
Sbjct: 66 LPGELVSNVLSRLPSKVLLLCKCVCKSWFDLITDPHFVSNYYVVYNSLQSQEEHLLVIRR 245
Query: 65 QHNFNSFDENVLNLPISLLLENS------LSTVPYDPYYRLKNENPHCPWL-FAGSCNGL 117
F+ L IS+L N+ +S+ +P Y ++ H W G CNG+
Sbjct: 246 PF-FSG-----LKTYISVLSWNTNDPKKHVSSDVLNPPYEYNSD--HKYWTEILGPCNGI 401
Query: 118 ICLCLDIDTSHGSRLCLWNPA-----TRTKSEFDLASQECFVF----AFGYDNLNGNYKV 168
L G+ L NP+ KS F + + F FG+D +YKV
Sbjct: 402 YFL-------EGNPNVLMNPSLGEFKALPKSHF-TSPHGTYTFTDYAGFGFDPKTNDYKV 557
Query: 169 IAF-DIKVKSGNARSV----VKVFSMRDNCWRNIQ-CFPVLPLYMFVSTQNGVYFSSTVN 222
+ D+ +K + R + +++S+ N WR + LP+ ++ S++ Y ++ +
Sbjct: 558 VVLKDLWLKETDEREIGYWSAELYSLNSNSWRKLDPSLLPLPIEIWGSSRVFTYANNCCH 737
Query: 223 WLALQDYFGLDYFHLNYSSITPEKYVILSLDLSTETYTQLLLPRGFNKVSRHQPKLAVLM 282
W + + S T + V+L+ D+ E++ ++ +P+ + L
Sbjct: 738 W----------WGFVEESDATQD--VVLAFDMVKESFRKIRVPKIRDSSDEKFGTLVPFE 881
Query: 283 DCLCFGH-----DYEETYFVIWQMKDFGVQSSWIQLFKITYQNFFSYYCDFARESK--WL 335
+ G E F +W MKD+ + SW++ + + F ++ W
Sbjct: 882 ESASIGFLVYPVRGTEKRFDVWVMKDYWDEGSWVKQYSVGPVQVIYKLVGFYGTNRFFWK 1061
Query: 336 DFLPLCLSKNGDTLILANNQE-NEAFIYNRRDDRVEKIGITNKNMLWFEAWDYVESLVST 394
D + + L+L +++ + +Y + D A Y ESLVS
Sbjct: 1062D--------SNERLVLYDSENTRDLQVYGKHDS--------------IRAAKYTESLVSL 1175
Query: 395 H 395
H
Sbjct: 1176H 1178
>TC223618
Length = 444
Score = 69.3 bits (168), Expect = 3e-12
Identities = 46/123 (37%), Positives = 60/123 (48%), Gaps = 5/123 (4%)
Frame = +1
Query: 84 LENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLCLDIDTSHGSRLCLWNPATRTKS 143
L N+LSTV + Y +KN+ H GSCNGL+C + G + LWNP+ R
Sbjct: 55 LFNNLSTVCDELNYPVKNKFRHDG--IVGSCNGLLCFAIK-----GDCVLLWNPSIRVSK 213
Query: 144 EFDLASQE----CFV-FAFGYDNLNGNYKVIAFDIKVKSGNARSVVKVFSMRDNCWRNIQ 198
+ CF F GYD++N +YKV+A VKV+SM N WR IQ
Sbjct: 214 KSPPLGNNWRPGCFTAFGLGYDHVNEDYKVVAVFCDPSEYFIECKVKVYSMATNSWRKIQ 393
Query: 199 CFP 201
FP
Sbjct: 394 DFP 402
>BI787361
Length = 423
Score = 63.5 bits (153), Expect = 2e-10
Identities = 43/125 (34%), Positives = 65/125 (51%), Gaps = 5/125 (4%)
Frame = +3
Query: 26 LMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQHNFNSFDENVLNLPISL--L 83
LMRFR V++ +N+LI DP F+++HL+ S +N ++++ + + V P S+ L
Sbjct: 6 LMRFRYVSETWNSLIFDPTFVKLHLERSPKNTHVLLEFQAIYDRDVGQQVGVAPCSIRRL 185
Query: 84 LENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLCLDIDTSHGSRLC---LWNPATR 140
+EN T+ D K+ N GSCNGL+C+ D C LWNPAT
Sbjct: 186 VENPSFTID-DCLTLFKHTNS-----IFGSCNGLVCMTKCFDVREFEEECQYRLWNPATG 347
Query: 141 TKSEF 145
SE+
Sbjct: 348 IMSEY 362
>TC216702 similar to UP|Q9LDP5 (Q9LDP5) FLORICAULA/LEAFY-like protein,
partial (5%)
Length = 1631
Score = 61.2 bits (147), Expect = 8e-10
Identities = 55/265 (20%), Positives = 121/265 (44%), Gaps = 18/265 (6%)
Frame = +3
Query: 132 LCLWNPATRT-----------KSEFDLASQECFVFAFGYDNLNGNYKV--IAFDIKVKSG 178
+ WNP+ R + D V FG+D+ +YK+ I++ + +
Sbjct: 30 IAFWNPSLRQHRILPYLPVPRRRHPDTTLFAARVCGFGFDHKTRDYKLVRISYFVDLHDR 209
Query: 179 NARSVVKVFSMRDNCWRNIQCFPVLPLYMFVSTQNGVYFSSTVNWLALQDYFGLDYFHLN 238
+ + VK++++R N W+ + P LP + + GV+ ++++W+ +
Sbjct: 210 SFDAQVKLYTLRANAWKTL---PSLPYALCCARTMGVFVGNSLHWVVTR----------- 347
Query: 239 YSSITPEKY-VILSLDLSTETYTQLLLPRGFNKVSRHQPKLAVLMDCLCFGHDYEETYFV 297
+ P++ +I++ DL+ + + +L LP + LA+L LC ++ +T
Sbjct: 348 --KLEPDQPDLIIAFDLTHDIFRELPLPDTGGVDGGFEIDLALLGGSLCMTVNFHKTRID 521
Query: 298 IWQMKDFGVQSSWIQLFKITYQNFFSYYCDFARESKWLDFL-PLCLSKNGDTLILANNQE 356
+W M+++ + SW ++F + + +RE + L + PL S +G+ ++L ++++
Sbjct: 522 VWVMREYNRRDSWCKVFTL----------EESREMRSLKCVRPLGYSSDGNKVLLEHDRK 671
Query: 357 NEAFIYNRRDDRVEKI---GITNKN 378
F Y+ V + G+ N N
Sbjct: 672 -RLFWYDLEKKEVALVKIQGLPNLN 743
>TC233794 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {Arabidopsis
thaliana;} , partial (9%)
Length = 449
Score = 56.6 bits (135), Expect = 2e-08
Identities = 43/129 (33%), Positives = 62/129 (47%), Gaps = 10/129 (7%)
Frame = +1
Query: 2 EDRHQSVLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMV 61
+++ + LP EL+ +IL LPVK L+RF+CV K + +LISDP F+ H +A +P +
Sbjct: 88 QNQSLTTLPQELIREILLRLPVKSLLRFKCVCKSFLSLISDPQFVISHYALAA-SPTHRL 264
Query: 62 IARQHNFNSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWL----------FA 111
I R H+F + S+ E+ T D Y P P L
Sbjct: 265 ILRSHDFYA---------QSIATESVFKTCSRDVVY-FPLPLPSIPCLRLDDFGIRPKIL 414
Query: 112 GSCNGLICL 120
GSC GL+ L
Sbjct: 415 GSCRGLVLL 441
>BG404995 similar to GP|22831068|dbj OJ1131_E05.32 {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 394
Score = 54.3 bits (129), Expect = 9e-08
Identities = 41/120 (34%), Positives = 61/120 (50%), Gaps = 8/120 (6%)
Frame = +1
Query: 9 LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNS---ARNPNLMVIA-- 63
LP E++T+ILS LPV+ L+RFR +K + +LI H +HL S A N +L++
Sbjct: 40 LPREVLTEILSRLPVRSLLRFRSTSKSWKSLIDSQHLNWLHLTRSLTLASNTSLILRVDS 219
Query: 64 --RQHNFNSFDENV-LNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICL 120
Q NF + D V LN P+ + NS++ + GSCNGL+C+
Sbjct: 220 DLYQTNFPTLDPPVSLNHPL-MCYSNSITLL--------------------GSCNGLLCI 336
>BE917614
Length = 388
Score = 53.1 bits (126), Expect = 2e-07
Identities = 30/81 (37%), Positives = 47/81 (57%)
Frame = -1
Query: 312 QLFKITYQNFFSYYCDFARESKWLDFLPLCLSKNGDTLILANNQENEAFIYNRRDDRVEK 371
+L ++Y NF E WL PLC+S+N D ++LAN +E ++NRRD+R++
Sbjct: 388 RLLNVSYLNF---QLSPTNELDWLPTTPLCISENDDMMLLANCVYDEFVLHNRRDNRIDS 218
Query: 372 IGITNKNMLWFEAWDYVESLV 392
IG + + ++DYV SLV
Sbjct: 217 IGSFDGKVPMC-SYDYVPSLV 158
>AI443156
Length = 414
Score = 51.6 bits (122), Expect = 6e-07
Identities = 39/129 (30%), Positives = 62/129 (47%)
Frame = +3
Query: 9 LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQHNF 68
LP E++T+ILS LPVK ++R R K++ ++I HFI HL S + +++ +
Sbjct: 90 LPVEVVTEILSRLPVKSVIRLRSTCKWWRSIIDSRHFILFHLNKSHTS---LILRHRSQL 260
Query: 69 NSFDENVLNLPISLLLENSLSTVPYDPYYRLKNENPHCPWLFAGSCNGLICLCLDIDTSH 128
S D L++ L P++ + L + L GS NGL+C+ D
Sbjct: 261 YSLD-----------LKSLLDPNPFELSHPLMCYSNSIKVL--GSSNGLLCISNVXDD-- 395
Query: 129 GSRLCLWNP 137
+ LWNP
Sbjct: 396 ---IALWNP 413
>TC213957 similar to GB|AAP37713.1|30725382|BT008354 At3g23880 {Arabidopsis
thaliana;} , partial (9%)
Length = 696
Score = 48.5 bits (114), Expect = 5e-06
Identities = 29/75 (38%), Positives = 42/75 (55%), Gaps = 1/75 (1%)
Frame = +3
Query: 1 MEDRHQSV-LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNL 59
ME SV LP EL+ +IL PV+ ++RF+CV K + +LISDP F L S + +
Sbjct: 6 MEKHTLSVTLPLELIREILLRSPVRSVLRFKCVCKSWLSLISDPQFTHFDLAASPTHRLI 185
Query: 60 MVIARQHNFNSFDEN 74
++ NFN + N
Sbjct: 186LISNYYDNFNYIESN 230
>AW395660
Length = 381
Score = 47.8 bits (112), Expect = 9e-06
Identities = 22/39 (56%), Positives = 31/39 (79%)
Frame = +3
Query: 9 LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQ 47
LP EL+ +ILS LPVK L++FRCV K + +LI DP+F++
Sbjct: 264 LPDELVVEILSRLPVKSLLQFRCVCKSWMSLIYDPYFMK 380
>BM143422
Length = 424
Score = 47.4 bits (111), Expect = 1e-05
Identities = 25/65 (38%), Positives = 37/65 (56%)
Frame = +1
Query: 5 HQSVLPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIAR 64
H LP +L+ IL LPVK + RF+CV K + +LISDP F H + P+ ++ R
Sbjct: 217 HTQTLPLDLIELILLKLPVKSVTRFKCVCKSWLSLISDPQFGFSHFDLALAVPSHRLLLR 396
Query: 65 QHNFN 69
+ F+
Sbjct: 397 SNEFS 411
>TC212036
Length = 501
Score = 43.1 bits (100), Expect = 2e-04
Identities = 23/57 (40%), Positives = 35/57 (61%)
Frame = -1
Query: 339 PLCLSKNGDTLILANNQENEAFIYNRRDDRVEKIGITNKNMLWFEAWDYVESLVSTH 395
PLC+S++ D ++L + +YNRR +R E++ KN F +DYV+SLVS H
Sbjct: 486 PLCMSQDEDVVLLTSYAGARFVLYNRRYNRSERME-HFKNKFSFYCYDYVQSLVSPH 319
>TC233972 weakly similar to GB|AAP37713.1|30725382|BT008354 At3g23880
{Arabidopsis thaliana;} , partial (10%)
Length = 631
Score = 41.6 bits (96), Expect = 6e-04
Identities = 23/55 (41%), Positives = 34/55 (61%), Gaps = 1/55 (1%)
Frame = -2
Query: 9 LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHL-KNSARNPNLMVI 62
LP E++ +IL LPVK L F+ V K +LIS+PHF + H +N+A L+ +
Sbjct: 540 LPQEIIIEILLRLPVKSLPSFKFVCKS*LSLISNPHFAKWHFERNAAHTEKLLFV 376
>TC216704 similar to UP|Q7PIN1 (Q7PIN1) ENSANGP00000023661 (Fragment),
partial (5%)
Length = 1232
Score = 39.3 bits (90), Expect = 0.003
Identities = 19/69 (27%), Positives = 37/69 (53%)
Frame = +2
Query: 248 VILSLDLSTETYTQLLLPRGFNKVSRHQPKLAVLMDCLCFGHDYEETYFVIWQMKDFGVQ 307
+I++ DL+ E +T+L LP + +A+L D LC ++ + +W M+++
Sbjct: 62 LIVAFDLTHEIFTELPLPDTGGVGGGFEIDVALLGDSLCMTVNFHNSKMDVWVMREYNRG 241
Query: 308 SSWIQLFKI 316
SW +LF +
Sbjct: 242 DSWCKLFTL 268
>TC219822 homologue to UP|CENB_HUMAN (P07199) Major centromere autoantigen B
(Centromere protein B) (CENP-B), partial (5%)
Length = 710
Score = 38.9 bits (89), Expect = 0.004
Identities = 20/63 (31%), Positives = 33/63 (51%)
Frame = +1
Query: 9 LPSELMTKILSLLPVKPLMRFRCVNKFYNTLISDPHFIQMHLKNSARNPNLMVIARQHNF 68
LP LM +IL + V ++ RCV K + +L+ DP F++ HL S + + +
Sbjct: 196 LPEGLMIEILVWIRVSNPLQLRCVCKRWKSLVVDPQFVKKHLHTSLSDITDLASNAMEDM 375
Query: 69 NSF 71
N+F
Sbjct: 376 NAF 384
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.325 0.139 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,870,082
Number of Sequences: 63676
Number of extensions: 406328
Number of successful extensions: 2804
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 2769
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2788
length of query: 395
length of database: 12,639,632
effective HSP length: 99
effective length of query: 296
effective length of database: 6,335,708
effective search space: 1875369568
effective search space used: 1875369568
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148651.11


