

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148607.13 + phase: 0
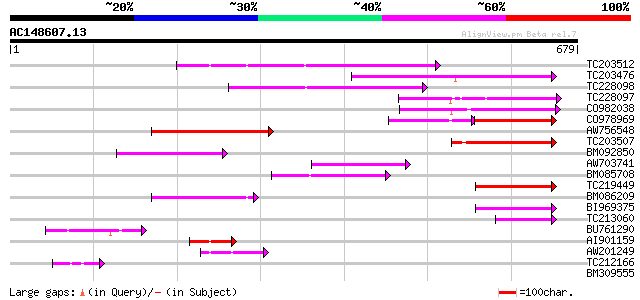
(679 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC203512 weakly similar to PIR|C86287|C86287 F9L1.24 protein - A... 196 2e-50
TC203476 weakly similar to PIR|C86287|C86287 F9L1.24 protein - A... 170 2e-42
TC228098 155 6e-38
TC228097 118 1e-26
CO982038 117 2e-26
CO978969 83 2e-23
AW756548 105 9e-23
TC203507 100 3e-21
BM092850 86 7e-17
AW703741 84 2e-16
BM085708 82 1e-15
TC219449 76 4e-14
BM086209 70 2e-12
BI969375 65 1e-10
TC213060 57 3e-08
BU761290 48 1e-05
AI901159 48 1e-05
AW201249 44 3e-04
TC212166 42 9e-04
BM309555 36 0.049
>TC203512 weakly similar to PIR|C86287|C86287 F9L1.24 protein - Arabidopsis
thaliana {Arabidopsis thaliana;} , partial (9%)
Length = 988
Score = 196 bits (499), Expect = 2e-50
Identities = 108/317 (34%), Positives = 179/317 (56%), Gaps = 1/317 (0%)
Frame = +3
Query: 200 GRVCLTSDCWTACSNEGYISLTAHYVNVNWKLESKILAFAHMEP-PHSGRDLALKVLEML 258
GRVCLT D WT+ + GY+ +T H+V+ +WKL+ +IL MEP P+S L+ V +
Sbjct: 36 GRVCLTLDVWTSSQSVGYVFITGHFVDSDWKLQRRILNVV-MEPYPNSDSALSHAVAVCI 212
Query: 259 DDWGIEKKIFSITLDNASANNSMANFLKEHLGLSNSLLLDGEFFHIRCSAHILNLIVQDG 318
DW E K+FSIT + + ++ N L+ L + N L+L+G+ C A L+ + D
Sbjct: 213 SDWNFEGKLFSITCGPSLSEVALGN-LRPLLFVKNPLILNGQLLIGNCIAQTLSSVANDL 389
Query: 319 LKVVSDALHKIRQSVAYVRVTESRTLLFSECVRIVGDIDTTIGLRLDCVTRWNSTFIMLQ 378
L V + KIR SV YV+ +ES F + + + + + L +D T+WN+T+ ML
Sbjct: 390 LSSVHLTVKKIRDSVKYVKTSESHEEKFLDLKQQL-QVPSERNLFIDDQTKWNTTYQMLV 566
Query: 379 SALVYRRAFYSLSLRDSNFKCCPTSEEWRRAEIMCEILKPFFTITNLISGSSYPTSNLYF 438
+A + F L D ++K P+ ++W+ E +C LKP F N+++ +++PT +F
Sbjct: 567 AASELQEVFSCLDTSDPDYKGAPSMQDWKLVETLCTYLKPLFDAANILTTATHPTVITFF 746
Query: 439 GEIWKIECLIRSYLTSEDLLIQKMAENMKVKFDKYWSDYNVVLAVGAVLDPTKKFNFLKF 498
E+WK++ + + SED I + + M+ K DKYW D ++VLA+ V+DP K ++F
Sbjct: 747 HEVWKLQLDLSRAVVSEDPFISNLTKPMQQKIDKYWKDCSLVLAIAVVMDPRFKMKLVEF 926
Query: 499 AYEKLDPLTSEEKLKKV 515
++ K+ + E +K V
Sbjct: 927 SFTKIYGEDAHEYVKIV 977
>TC203476 weakly similar to PIR|C86287|C86287 F9L1.24 protein - Arabidopsis
thaliana {Arabidopsis thaliana;} , partial (19%)
Length = 1227
Score = 170 bits (430), Expect = 2e-42
Identities = 85/254 (33%), Positives = 147/254 (57%), Gaps = 8/254 (3%)
Frame = +1
Query: 410 EIMCEILKPFFTITNLISGSSYPTSNLYFGEIWKIECLIRSYLTSEDLLIQKMAENMKVK 469
E +C LKP F N+++ +++ T +F E+WK++ + + +ED I + + M+ K
Sbjct: 160 ETLCTYLKPLFDAANILTTTTHTTVITFFHEVWKLQLDLSRAIVNEDPFISNLTKPMQQK 339
Query: 470 FDKYWSDYNVVLAVGAVLDPTKKFNFLKFAYEKLDPLTSEEKLKKVKMTLGKLFSEYIKN 529
DKYW D +VVLA+ V+DP K ++F++ K+ + E +K V + +LF EY+
Sbjct: 340 IDKYWKDCSVVLAIAVVMDPRFKMKLVEFSFTKIYGEDAHEYVKIVDDGIHELFHEYVTL 519
Query: 530 GIP--------SNLSSSQVQPSYGGGTRITSSSYDEFEEYESQSSNSTGKSELDTYLDEL 581
+P N S GGT + + +F+ Y ++S+ KSELD YL+E
Sbjct: 520 PLPLTPAYAEEGNAGSHPRAGESPGGTLMPDNGLTDFDVYIMETSSHQMKSELDQYLEES 699
Query: 582 RMPLSQEFDVLAFWKERSRRSPNLARMACDILSIPITTVASESAFSIGARVVNRYRSSMK 641
+P +FDVL +WK + P L++MA DILS+P+++V ES F + +++YRSS++
Sbjct: 700 LLPRVPDFDVLGWWKLNKLKYPTLSKMARDILSVPVSSVPPESVFDTKVKEMDQYRSSLR 879
Query: 642 DDSVQALLCARSWL 655
++V+A++CA+ W+
Sbjct: 880 PETVEAIVCAKDWM 921
>TC228098
Length = 720
Score = 155 bits (392), Expect = 6e-38
Identities = 83/238 (34%), Positives = 133/238 (55%)
Frame = +2
Query: 263 IEKKIFSITLDNASANNSMANFLKEHLGLSNSLLLDGEFFHIRCSAHILNLIVQDGLKVV 322
I+ K+F++TLD+ S ++ + +KE + L + +R +AH++N I QD + +
Sbjct: 5 IDHKLFAVTLDDCSIDDDITLRIKERVSEKRPFLSTRQLLDVRSAAHLVNSIAQDAMDAL 184
Query: 323 SDALHKIRQSVAYVRVTESRTLLFSECVRIVGDIDTTIGLRLDCVTRWNSTFIMLQSALV 382
+ + KIR+S+ Y+R ++ F+E + I+T L LD +W ST++ML++AL
Sbjct: 185 HEVIQKIRESIRYIRSSQVVQGKFNEIAQRAR-INTQKFLFLDFPVQWKSTYLMLETALE 361
Query: 383 YRRAFYSLSLRDSNFKCCPTSEEWRRAEIMCEILKPFFTITNLISGSSYPTSNLYFGEIW 442
YR AF D ++ T EEW A + LK ITN+ SG+ +PT+N+YF EI
Sbjct: 362 YRSAFSLFQEHDPSYSASLTDEEWEWASSVTGYLKLLVEITNVFSGNKFPTANIYFPEIC 541
Query: 443 KIECLIRSYLTSEDLLIQKMAENMKVKFDKYWSDYNVVLAVGAVLDPTKKFNFLKFAY 500
+ + + S D + MA MK KFD+YWS ++ LAV AVLDP K +++ Y
Sbjct: 542 DVHMQLIDWCRSSDDFLSPMALKMKAKFDRYWSKCSLALAVAAVLDPRFKMKLVEYYY 715
>TC228097
Length = 832
Score = 118 bits (295), Expect = 1e-26
Identities = 74/201 (36%), Positives = 111/201 (54%), Gaps = 6/201 (2%)
Frame = +2
Query: 466 MKVKFDKYWSDYNVVLAVGAVLDPTKKFNFLKFAYEKLDPLTSEEKLKKVKMTLGKLFSE 525
MK KFD+YWS ++ LAV AVLDP K +++ + + T+ E +K+V + +LF+
Sbjct: 2 MKDKFDRYWSKCSLPLAVAAVLDPRFKMKLVEYYFSLIYGSTALEHIKEVSDGIKELFNA 181
Query: 526 Y------IKNGIPSNLSSSQVQPSYGGGTRITSSSYDEFEEYESQSSNSTGKSELDTYLD 579
Y I G S L S + PS +R +D F SQ + S+LD YL+
Sbjct: 182 YSICSTMIDQG--SALPGSSL-PSTSCSSRDRLKGFDRFLHETSQGQSMI--SDLDKYLE 346
Query: 580 ELRMPLSQEFDVLAFWKERSRRSPNLARMACDILSIPITTVASESAFSIGARVVNRYRSS 639
E P + +F++L +WK R P L+ MA D+L P++T+A E AFS G RV++ RSS
Sbjct: 347 EPIFPRNSDFNILNWWKVHMPRYPILSMMARDVLGTPMSTMAPELAFSTGGRVLDSSRSS 526
Query: 640 MKDDSVQALLCARSWLHGFEG 660
+ D+ +AL+C + WL G
Sbjct: 527 LNPDTREALICTQDWLQNESG 589
>CO982038
Length = 675
Score = 117 bits (293), Expect = 2e-26
Identities = 73/196 (37%), Positives = 110/196 (55%), Gaps = 4/196 (2%)
Frame = -1
Query: 468 VKFDKYWSDYNVVLAVGAVLDPTKKFNFLKFAYEKLDPLTSEEKLKKVKMTLGKLFSEYI 527
+KFDKYW + V+ V VLD K L++ YEKL S +++ ++ L S+Y
Sbjct: 675 IKFDKYWGVIHNVMGVATVLDLRYKMELLEYYYEKLYEHDSFTQVRGIQQLCYDLVSDYQ 496
Query: 528 ----KNGIPSNLSSSQVQPSYGGGTRITSSSYDEFEEYESQSSNSTGKSELDTYLDELRM 583
K+ SN+ G S YD F + ++ +S KSELD YL E +
Sbjct: 495 MKMNKDSFGSNVGDVTGSEVVGDAL----SEYDRFII*KKRARSSYVKSELDNYL*EDVL 328
Query: 584 PLSQEFDVLAFWKERSRRSPNLARMACDILSIPITTVASESAFSIGARVVNRYRSSMKDD 643
P + +FD+L +WK + P L +A DIL+I ++TVASESAFS G +V++ +RS ++
Sbjct: 327 PRAVDFDILMWWKFNGVKYPTLQAIAKDILAILVSTVASESAFSTGGQVLSPHRSRLQWT 148
Query: 644 SVQALLCARSWLHGFE 659
+++AL+CARSWL E
Sbjct: 147 TLEALMCARSWLWSAE 100
>CO978969
Length = 747
Score = 82.8 bits (203), Expect(2) = 2e-23
Identities = 40/100 (40%), Positives = 66/100 (66%), Gaps = 1/100 (1%)
Frame = -3
Query: 557 EFEEYESQ-SSNSTGKSELDTYLDELRMPLSQEFDVLAFWKERSRRSPNLARMACDILSI 615
+FE Y S + N K+ELD YL+E P QEFD+L++W+ + P L+R+A DIL +
Sbjct: 406 DFEFYISDFTCNQQFKTELDEYLEEPLEPRVQEFDILSWWRINGLKYPTLSRLASDILCM 227
Query: 616 PITTVASESAFSIGARVVNRYRSSMKDDSVQALLCARSWL 655
PI+T+++++ F R ++ YRSS+ +++AL+C + WL
Sbjct: 226 PISTLSADAVFDTEIRKMDSYRSSLDSLTLEALICTKDWL 107
Score = 45.1 bits (105), Expect(2) = 2e-23
Identities = 28/106 (26%), Positives = 50/106 (46%)
Frame = -1
Query: 454 SEDLLIQKMAENMKVKFDKYWSDYNVVLAVGAVLDPTKKFNFLKFAYEKLDPLTSEEKLK 513
S+D + ++ KFD+YW + VLAV V+DP K ++ + K+ +++ ++
Sbjct: 747 SQDPFCSHLITPLQEKFDQYWRESCFVLAVAVVMDPRYKMKLVESTFAKIFGENADQWIR 568
Query: 514 KVKMTLGKLFSEYIKNGIPSNLSSSQVQPSYGGGTRITSSSYDEFE 559
V+ L +LF EY I L + G I + + EF+
Sbjct: 567 IVEDGLHELFLEY---NIIQVLPFTATNDDEGNEAMIKTEPFQEFD 439
>AW756548
Length = 445
Score = 105 bits (261), Expect = 9e-23
Identities = 46/146 (31%), Positives = 92/146 (62%)
Frame = +3
Query: 171 VSRNTIAKEILNVYRDEKQKLKSQLAQIRGRVCLTSDCWTACSNEGYISLTAHYVNVNWK 230
VS N + + + +Y E++K+ L ++ G++ L++D W A + Y+ LT++Y++ +W+
Sbjct: 3 VSVNKVEADCIEIYERERKKVNEMLDKLPGKISLSADVWNAVDDAEYLCLTSNYIDESWQ 182
Query: 231 LESKILAFAHMEPPHSGRDLALKVLEMLDDWGIEKKIFSITLDNASANNSMANFLKEHLG 290
L +IL F ++P H+ ++ ++ L DW I++K+FS+ LD+ S ++ +A + E L
Sbjct: 183 LRRRILNFIRIDPSHTEDMVSEAIMTCLMDWDIDRKLFSMILDSCSTSDKIAVRIGERLL 362
Query: 291 LSNSLLLDGEFFHIRCSAHILNLIVQ 316
+ L +G+ F IRC+A+++N +VQ
Sbjct: 363 QNRFLYCNGQLFDIRCAANVINTMVQ 440
>TC203507
Length = 709
Score = 100 bits (248), Expect = 3e-21
Identities = 48/126 (38%), Positives = 80/126 (63%)
Frame = +3
Query: 530 GIPSNLSSSQVQPSYGGGTRITSSSYDEFEEYESQSSNSTGKSELDTYLDELRMPLSQEF 589
G P N + P GGT ++ + +F+ Y ++SN KSELD YL+E +P +F
Sbjct: 6 GNPGNHPKTGGSP---GGTMMSDNGLTDFDVYIMETSNHQMKSELDQYLEESLLPRVPDF 176
Query: 590 DVLAFWKERSRRSPNLARMACDILSIPITTVASESAFSIGARVVNRYRSSMKDDSVQALL 649
DVL +WK + P L++MA DILS+P++++ ES F + +++YRSS++ ++V+AL+
Sbjct: 177 DVLGWWKLNKLKYPTLSKMARDILSVPVSSLPPESVFDTKVKEMDQYRSSLRPETVEALV 356
Query: 650 CARSWL 655
CA+ W+
Sbjct: 357 CAKDWM 374
>BM092850
Length = 421
Score = 85.5 bits (210), Expect = 7e-17
Identities = 40/134 (29%), Positives = 78/134 (57%)
Frame = +1
Query: 128 KFDPMANREFLARIMITHGAPFNMVEWKVFREYQKFLNDDCVFVSRNTIAKEILNVYRDE 187
KFD ++ LAR++I HG P ++VE F+ + K L F +++ +++YR E
Sbjct: 19 KFDQERSQLDLARMIILHGYPLSLVEQVGFKVFVKNLQPLFEFTPNSSVEISCIDIYRRE 198
Query: 188 KQKLKSQLAQIRGRVCLTSDCWTACSNEGYISLTAHYVNVNWKLESKILAFAHMEPPHSG 247
K+K+ + ++ GR+ L+ + W++ N Y+ L+AHY++ W L+ K+L F ++ H+
Sbjct: 199 KEKVFDMINRLHGRINLSIETWSSTENSLYLCLSAHYIDEEWTLQKKLLNFVTLDSSHTE 378
Query: 248 RDLALKVLEMLDDW 261
L +++ L++W
Sbjct: 379 DLLPEVIIKCLNEW 420
>AW703741
Length = 412
Score = 84.0 bits (206), Expect = 2e-16
Identities = 40/120 (33%), Positives = 68/120 (56%), Gaps = 1/120 (0%)
Frame = +3
Query: 362 LRLDCVTRWNSTFIMLQSALVYRRAFYSLSLRDSNFKCCPTS-EEWRRAEIMCEILKPFF 420
L +D WN ++ ML +A + F L D ++K P S ++W+ EI+C LKP F
Sbjct: 51 LFIDDQIHWNRSYQMLVAASELKEVFSCLDTSDPDYKGAPPSMQDWKLVEILCSYLKPLF 230
Query: 421 TITNLISGSSYPTSNLYFGEIWKIECLIRSYLTSEDLLIQKMAENMKVKFDKYWSDYNVV 480
N+++ +++PT+ +F E+WK++ +TSED I + + M K DKYW + ++V
Sbjct: 231 DAANILTTTTHPTTIAFFHEVWKLQLDAARAVTSEDPFISNLNKIMSEKIDKYWRECSLV 410
>BM085708
Length = 428
Score = 81.6 bits (200), Expect = 1e-15
Identities = 44/143 (30%), Positives = 77/143 (53%)
Frame = +2
Query: 314 IVQDGLKVVSDALHKIRQSVAYVRVTESRTLLFSECVRIVGDIDTTIGLRLDCVTRWNST 373
+VQ L V + ++KIRQ++ Y++ ++ F+ + VG + GL LD ++WNST
Sbjct: 2 MVQHALGTVREIVNKIRQTIGYIKSSQIILAKFNVMAKEVGILSQK-GLFLDNPSQWNST 178
Query: 374 FIMLQSALVYRRAFYSLSLRDSNFKCCPTSEEWRRAEIMCEILKPFFTITNLISGSSYPT 433
+ ML AL ++ L D+ +K C + EW R + LK F + N+ + S YPT
Sbjct: 179 YSMLVVALEFKDVLILLQENDTAYKVCLSEVEWERVTAVTSYLKLFVEVINVFTKSKYPT 358
Query: 434 SNLYFGEIWKIECLIRSYLTSED 456
+N+YF E+ ++ + + + D
Sbjct: 359 ANIYFPELCDVKLHLIEWSKNSD 427
>TC219449
Length = 702
Score = 76.3 bits (186), Expect = 4e-14
Identities = 38/99 (38%), Positives = 67/99 (67%), Gaps = 1/99 (1%)
Frame = +3
Query: 558 FEEYESQSSNSTG-KSELDTYLDELRMPLSQEFDVLAFWKERSRRSPNLARMACDILSIP 616
F+++ +++ G KS+LD YL+E P + +F++L +W+ + R P L+ MA ++L IP
Sbjct: 156 FDKFLHETTQGEGTKSDLDKYLEEPLFPRNVDFNILNWWRVHTPRYPVLSMMARNVLGIP 335
Query: 617 ITTVASESAFSIGARVVNRYRSSMKDDSVQALLCARSWL 655
++ VA E AF+ ARV++R SS+ +VQAL+C++ W+
Sbjct: 336 MSKVAPELAFNHSARVLDRDWSSLNPATVQALVCSQDWI 452
>BM086209
Length = 389
Score = 70.5 bits (171), Expect = 2e-12
Identities = 38/128 (29%), Positives = 68/128 (52%)
Frame = +1
Query: 171 VSRNTIAKEILNVYRDEKQKLKSQLAQIRGRVCLTSDCWTACSNEGYISLTAHYVNVNWK 230
+S N I + + +Y EKQ L + + I GRV LT D WT+ Y+ L H+++ +W
Sbjct: 7 LSFNAIQGDCVTMYLREKQNLLNLIDGIPGRVNLTLDLWTSNQTSAYVFLRGHFIDGDWN 186
Query: 231 LESKILAFAHMEPPHSGRDLALKVLEMLDDWGIEKKIFSITLDNASANNSMANFLKEHLG 290
L IL + P S L ++ L DW ++ ++F++ LD +N ++ L+ L
Sbjct: 187 LHHPILNVFMVPFPDSDGSLNQTIVNCLSDWHLKGRLFTLALDKFFSNETLGK-LRSLLS 363
Query: 291 LSNSLLLD 298
++N ++L+
Sbjct: 364 VNNPVILN 387
>BI969375
Length = 510
Score = 65.1 bits (157), Expect = 1e-10
Identities = 32/98 (32%), Positives = 59/98 (59%)
Frame = -3
Query: 558 FEEYESQSSNSTGKSELDTYLDELRMPLSQEFDVLAFWKERSRRSPNLARMACDILSIPI 617
F+ Y + + K E+D L+E +P DVLA+W+ + + L++M DILS P+
Sbjct: 412 FDAYIMGTPSHHTKYEIDQCLEESLLPRVPHSDVLAWWEINTIKCLTLSKMGRDILSTPV 233
Query: 618 TTVASESAFSIGARVVNRYRSSMKDDSVQALLCARSWL 655
T A +S ++ ++ YRSS++ ++++AL+CA+ W+
Sbjct: 232 FTGALDSVCYSKSKEIDEYRSSLRPETLEALVCAKGWM 119
>TC213060
Length = 499
Score = 56.6 bits (135), Expect = 3e-08
Identities = 29/74 (39%), Positives = 41/74 (55%)
Frame = +2
Query: 582 RMPLSQEFDVLAFWKERSRRSPNLARMACDILSIPITTVASESAFSIGARVVNRYRSSMK 641
R+ L DVL +WK S R P L+ MA D L++ T+V E F +++ R M
Sbjct: 107 RLQLQIPADVLEWWKVNSTRYPRLSVMARDFLAVQATSVVPEELFCGKGDEIDKQRICMP 286
Query: 642 DDSVQALLCARSWL 655
DS QA+LC +SW+
Sbjct: 287 HDSTQAILCIKSWI 328
>BU761290
Length = 441
Score = 48.1 bits (113), Expect = 1e-05
Identities = 41/130 (31%), Positives = 63/130 (47%), Gaps = 9/130 (6%)
Frame = +2
Query: 44 DKDTGKVKSQGKA--LTSDVWLYLVKVGIVDGVEKCRCKACHKLLTCESGSGTSHLKRHV 101
+ + V++QG L S +W + K+ V+G++K CK C KLL +S +GT HL +H
Sbjct: 41 EDEVNNVEAQGGICRLKSKIWQHFKKIK-VNGLDKTECKYCKKLLGGKSKNGTKHLWQHN 217
Query: 102 RSC--SKTIKNHDVGEMMID---VEGK--LRKKKFDPMANREFLARIMITHGAPFNMVEW 154
C K G+ + V+GK L +D RE LA+ +I H P ++ +
Sbjct: 218 EICVQYKIFMRGMKGQAFLTPKVVQGKQELGAGTYDA---REQLAKAIIMHEYPLSIGDR 388
Query: 155 KVFREYQKFL 164
FR Y L
Sbjct: 389 LGFRRYSAAL 418
>AI901159
Length = 388
Score = 48.1 bits (113), Expect = 1e-05
Identities = 25/57 (43%), Positives = 36/57 (62%), Gaps = 1/57 (1%)
Frame = +1
Query: 216 GYISLTAHYVNVNWKLESKILAFAHMEP-PHSGRDLALKVLEMLDDWGIEKKIFSIT 271
GY+ +T H+V+ +WKL+ +IL MEP P+S L+ V + DW E K+FSIT
Sbjct: 199 GYVFITGHFVDSDWKLQRRILNVV-MEPYPNSDSALSHAVAVCISDWNCEGKLFSIT 366
>AW201249
Length = 405
Score = 43.5 bits (101), Expect = 3e-04
Identities = 31/82 (37%), Positives = 46/82 (55%), Gaps = 1/82 (1%)
Frame = +1
Query: 229 WKLESKILAFAHMEPPHSGRDLALKVLEMLDDWGIEKKIFSITLDNASANNSMANFLKEH 288
++L KIL F + H G + + L + GI+K I IT+DNASAN+ ++L
Sbjct: 19 FRLHKKILKFC-LIGDHKGETTWIALENCLKE*GIQK-ICCITVDNASANSVAISYLSRG 192
Query: 289 LGLSNS-LLLDGEFFHIRCSAH 309
L + N LL+ EF H++ SAH
Sbjct: 193 LSV*NDHTLLNEEFIHMQRSAH 258
>TC212166
Length = 653
Score = 42.0 bits (97), Expect = 9e-04
Identities = 24/62 (38%), Positives = 34/62 (54%)
Frame = +1
Query: 52 SQGKALTSDVWLYLVKVGIVDGVEKCRCKACHKLLTCESGSGTSHLKRHVRSCSKTIKNH 111
S+ +TS VW + ++ +G K C+ C KLL + GTSHLKRH+ C K K
Sbjct: 307 SKRPKMTSKVWEEMQRIQTTEG-SKVLCRHCGKLL--QDNCGTSHLKRHLIICPKRPKPL 477
Query: 112 DV 113
D+
Sbjct: 478 DI 483
>BM309555
Length = 435
Score = 36.2 bits (82), Expect = 0.049
Identities = 26/107 (24%), Positives = 42/107 (38%), Gaps = 5/107 (4%)
Frame = +1
Query: 11 AREIPPVALIDQNAQEVSSDPKDKKRKGKAKAKDKDTGKVKSQGKALTSDVWLYLVKVGI 70
A IP +++ S +P + + K + K S VW + +
Sbjct: 100 ASNIPSYEIVNAETPLNSEEPTPETQPSKRRKKK--------------SIVWEHFTIETV 237
Query: 71 VDGVEKCRCKACHKLLTCESGS---GTSHLKRHVR--SCSKTIKNHD 112
G + CK C + +GS GTSHLKRH+ +C ++ D
Sbjct: 238 SPGCRRACCKQCKQSFAYSTGSKVAGTSHLKRHIAKGTCPALLRGQD 378
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,033,982
Number of Sequences: 63676
Number of extensions: 436683
Number of successful extensions: 2368
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 2339
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2359
length of query: 679
length of database: 12,639,632
effective HSP length: 104
effective length of query: 575
effective length of database: 6,017,328
effective search space: 3459963600
effective search space used: 3459963600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148607.13


