

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148607.1 + phase: 0 /pseudo
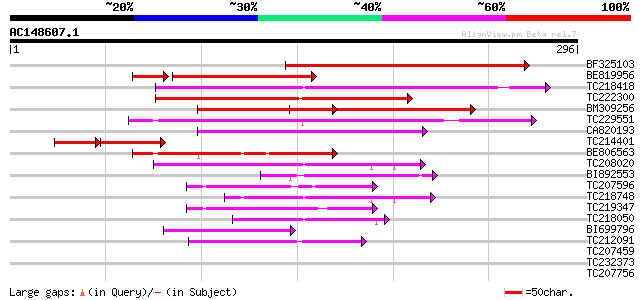
(296 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BF325103 182 2e-46
BE819956 similar to GP|20522008|db pleiotropic drug resistance l... 118 6e-32
TC218418 homologue to UP|Q6X4V5 (Q6X4V5) ABC transporter, partia... 100 8e-22
TC222300 similar to GB|AAP37786.1|30725528|BT008427 At1g51560 {A... 100 1e-21
BM309256 similar to PIR|T02644|T0 ABC-type transport protein hom... 99 2e-21
TC229551 weakly similar to UP|Q6X4V5 (Q6X4V5) ABC transporter, p... 92 3e-19
CA820193 similar to PIR|T04229|T04 ABC-type transport protein F1... 69 3e-12
TC214401 weakly similar to UP|Q949G3 (Q949G3) ABC1 protein, part... 46 4e-12
BE806563 similar to GP|11994752|db ABC transporter-like protein ... 63 1e-10
TC208020 similar to UP|Q76CU2 (Q76CU2) PDR-type ABC transporter ... 62 4e-10
BI892553 57 1e-08
TC207596 similar to UP|Q7GBY1 (Q7GBY1) Tonoplast ABC transporter... 47 8e-06
TC218748 similar to UP|Q76CU1 (Q76CU1) PDR-type ABC transporter ... 45 3e-05
TC219347 similar to UP|Q9LF78 (Q9LF78) Mitochondrial half-ABC tr... 45 5e-05
TC218050 weakly similar to UP|Q7PC82 (Q7PC82) PDR14 ABC transpor... 44 1e-04
BI699796 similar to PIR|G85167|G85 ABC transporter like protein ... 43 2e-04
TC212091 similar to UP|Q9LFG8 (Q9LFG8) ABC transporter-like prot... 42 3e-04
TC207459 similar to UP|Q94IH6 (Q94IH6) CjMDR1, partial (30%) 40 0.001
TC232373 weakly similar to UP|AB10_MOUSE (Q9JI39) ATP-binding ca... 39 0.004
TC207756 similar to UP|Q8RY46 (Q8RY46) Transporter associated wi... 37 0.011
>BF325103
Length = 393
Score = 182 bits (462), Expect = 2e-46
Identities = 81/127 (63%), Positives = 105/127 (81%)
Frame = -2
Query: 145 MRQYVHILKGTVVISLLQPPPETYNLFDDIILLSDSHIIYQGPREHVLEFFESIGFKCPN 204
+RQ VH++ T++ISLLQP PET++LFDDIILLS+ HIIYQGPRE+VL+FFES+GFKCP
Sbjct: 389 LRQLVHVMDVTMIISLLQPAPETFDLFDDIILLSEGHIIYQGPRENVLDFFESVGFKCPE 210
Query: 205 RKGVADFLQEVTSRKDQEQYWEHKDRPYRFITAEEFSEAFQTFHVGRRLGDELGTEFDKS 264
RKG+ADFLQEVTSRKDQEQYW +D+PYR+++ EF F F +G++L EL +D++
Sbjct: 209 RKGIADFLQEVTSRKDQEQYWFARDKPYRYVSVPEFVAHFNNFGIGQQLSQELKVPYDRA 30
Query: 265 NSHPAAL 271
+HPAAL
Sbjct: 29 KTHPAAL 9
>BE819956 similar to GP|20522008|db pleiotropic drug resistance like protein
{Nicotiana tabacum}, partial (6%)
Length = 673
Score = 118 bits (295), Expect(2) = 6e-32
Identities = 58/75 (77%), Positives = 67/75 (89%)
Frame = -1
Query: 86 GLEICADTVVGNAMIRAISGGQKKRLTTGEMLVGPTKALFMDEISTGLDSSTTFQIVNSM 145
GLE+CADT+VGNAM+R ISGGQ+KR+TT EMLVG A FMDEISTGLDSSTT+QI+NS+
Sbjct: 445 GLEVCADTIVGNAMLRGISGGQRKRVTTWEMLVGLANA*FMDEISTGLDSSTTYQILNSL 266
Query: 146 RQYVHILKGTVVISL 160
+Q VHILKGT VISL
Sbjct: 265 KQCVHILKGTTVISL 221
Score = 37.0 bits (84), Expect(2) = 6e-32
Identities = 16/19 (84%), Positives = 19/19 (99%)
Frame = -2
Query: 65 KAVATEGQKENLITDYVLR 83
KA+ATEG+KENL+TDYVLR
Sbjct: 501 KALATEGEKENLMTDYVLR 445
>TC218418 homologue to UP|Q6X4V5 (Q6X4V5) ABC transporter, partial (49%)
Length = 1220
Score = 100 bits (249), Expect = 8e-22
Identities = 63/207 (30%), Positives = 107/207 (51%), Gaps = 1/207 (0%)
Frame = +1
Query: 77 ITDYVLRVLGLEICADTVVGNAMIRAISGGQKKRLTTGEMLVGPTKALFMDEISTGLDSS 136
+ + + +GL+ CADTV+GN +R ISGG+K+R++ ++ + LF+DE ++GLDS+
Sbjct: 592 LVESTIVAMGLQDCADTVIGNWHLRGISGGEKRRVSIALEILMRPRLLFLDEPTSGLDSA 771
Query: 137 TTFQIVNSMRQYVHILKGTVVISLLQPPPETYNLFDDIILLSDSHIIYQGPREHVLEFFE 196
+ F + ++R + TV+ S+ QP E + LFD + LLS +Y G EFF
Sbjct: 772 SAFFVTQTLRALARDGR-TVIASIHQPSSEVFELFDQLYLLSSGKTVYFGQASEAYEFFA 948
Query: 197 SIGFKCPNRKGVAD-FLQEVTSRKDQEQYWEHKDRPYRFITAEEFSEAFQTFHVGRRLGD 255
GF CP + +D FL+ + S D+ + RF +++ + T R L D
Sbjct: 949 QAGFPCPALRNPSDHFLRCINSDFDKVKATLKGSMKLRFEGSDDPLDRITTAEAIRTLID 1128
Query: 256 ELGTEFDKSNSHPAALTTKKYGVGKIE 282
F +++ H A K + K++
Sbjct: 1129-----FYRTSQHSYAARQKVDEISKVK 1194
>TC222300 similar to GB|AAP37786.1|30725528|BT008427 At1g51560 {Arabidopsis
thaliana;} , partial (37%)
Length = 801
Score = 100 bits (248), Expect = 1e-21
Identities = 50/134 (37%), Positives = 81/134 (60%)
Frame = +1
Query: 77 ITDYVLRVLGLEICADTVVGNAMIRAISGGQKKRLTTGEMLVGPTKALFMDEISTGLDSS 136
ITD + +GL+ C D ++G +R ISGG+KKRL+ ++ + LF+DE ++GLDS+
Sbjct: 4 ITDATIIEMGLQDCDDRLIGKWHLRGISGGEKKRLSIAL*ILTRPRLLFLDEPTSGLDSA 183
Query: 137 TTFQIVNSMRQYVHILKGTVVISLLQPPPETYNLFDDIILLSDSHIIYQGPREHVLEFFE 196
+ F +V ++R + TV+ S+ QP E + L+DD+ LLS +Y G + +EFF+
Sbjct: 184 SAFFVVQTLRNVARD-ERTVIYSIHQPDSEVFALYDDLFLLSGGETVYFGEAKSAIEFFD 360
Query: 197 SIGFKCPNRKGVAD 210
GF CP ++ D
Sbjct: 361 EAGFPCPRKRNPYD 402
>BM309256 similar to PIR|T02644|T0 ABC-type transport protein homolog
F12C20.5 - Arabidopsis thaliana, partial (9%)
Length = 441
Score = 99.0 bits (245), Expect = 2e-21
Identities = 48/97 (49%), Positives = 63/97 (64%)
Frame = +2
Query: 147 QYVHILKGTVVISLLQPPPETYNLFDDIILLSDSHIIYQGPREHVLEFFESIGFKCPNRK 206
Q VH++ G + N F +ILL + I+YQGPRE ++FF+ +GF CP RK
Sbjct: 152 QPVHLM-GPQLYPFFNQLQRPMNCFLYVILLCEGQIVYQGPREAAVDFFKQMGFSCPERK 328
Query: 207 GVADFLQEVTSRKDQEQYWEHKDRPYRFITAEEFSEA 243
VADFLQEVTS+KDQEQ W DRPYR++ +F+EA
Sbjct: 329 NVADFLQEVTSKKDQEQDWSVPDRPYRYVPVGKFAEA 439
Score = 92.8 bits (229), Expect = 2e-19
Identities = 42/73 (57%), Positives = 54/73 (73%)
Frame = +1
Query: 99 MIRAISGGQKKRLTTGEMLVGPTKALFMDEISTGLDSSTTFQIVNSMRQYVHILKGTVVI 158
++ + GG KRLTTGE+L+GP + LFMDEISTGLD STT+QI+ ++ L GT ++
Sbjct: 4 VVIVLPGGLHKRLTTGELLIGPARVLFMDEISTGLDRSTTYQIIRYLKHSTRALDGTTIV 183
Query: 159 SLLQPPPETYNLF 171
SLLQP PETY LF
Sbjct: 184 SLLQPAPETYELF 222
>TC229551 weakly similar to UP|Q6X4V5 (Q6X4V5) ABC transporter, partial (34%)
Length = 1436
Score = 92.0 bits (227), Expect = 3e-19
Identities = 60/215 (27%), Positives = 105/215 (47%), Gaps = 2/215 (0%)
Frame = +3
Query: 63 YMKAVATEGQKENLITDYVLRVLGLEICADTVVGNAMIRAISGGQKKRLTTGEMLVGPTK 122
+ ++ E +KE D LR +GL+ +T VG + +SGGQ++RL+ ++ K
Sbjct: 333 FPNTMSVEEKKER--ADMTLREMGLQDAINTRVGGWNCKGLSGGQRRRLSICIEILTHPK 506
Query: 123 ALFMDEISTGLDSSTTFQIVNSMRQYVHI--LKGTVVISLLQPPPETYNLFDDIILLSDS 180
LF+DE ++GLDS+ ++ +++ + + ++ T+V S+ QP E + LF D+ LLS
Sbjct: 507 LLFLDEPTSGLDSAASYYVMSGIANLIQRDGIQRTIVASVHQPSSEVFQLFHDLFLLSSG 686
Query: 181 HIIYQGPREHVLEFFESIGFKCPNRKGVADFLQEVTSRKDQEQYWEHKDRPYRFITAEEF 240
+Y GP +FF S GF CP +D + ++ + E IT EE
Sbjct: 687 ETVYFGPASDANQFFASNGFPCPPLYNPSDHYLRIINKDFNQDADEG-------ITTEEA 845
Query: 241 SEAFQTFHVGRRLGDELGTEFDKSNSHPAALTTKK 275
++ + + + +E KS + A KK
Sbjct: 846 TKILVNSYKSSEFSNHVQSEIAKSETDFGACGKKK 950
>CA820193 similar to PIR|T04229|T04 ABC-type transport protein F14M19.30 -
Arabidopsis thaliana, partial (23%)
Length = 423
Score = 68.6 bits (166), Expect = 3e-12
Identities = 33/120 (27%), Positives = 67/120 (55%)
Frame = +1
Query: 99 MIRAISGGQKKRLTTGEMLVGPTKALFMDEISTGLDSSTTFQIVNSMRQYVHILKGTVVI 158
+ R +SGG+++R++ G L+ L +DE ++GLDS++ F+++ ++Q T+++
Sbjct: 64 LARGLSGGERRRVSIGLCLLHDPAVLLLDEPTSGLDSTSAFKVMRILKQTCVSRNRTIIL 243
Query: 159 SLLQPPPETYNLFDDIILLSDSHIIYQGPREHVLEFFESIGFKCPNRKGVADFLQEVTSR 218
S+ QP + D I+LLS +++ G + F S GF P++ ++ E+ S+
Sbjct: 244 SIHQPSFKILACIDRILLLSKGQVVHHGSVATLQAFLHSNGFTVPHQLNALEYAMEILSQ 423
>TC214401 weakly similar to UP|Q949G3 (Q949G3) ABC1 protein, partial (18%)
Length = 1177
Score = 46.2 bits (108), Expect(2) = 4e-12
Identities = 21/34 (61%), Positives = 25/34 (72%)
Frame = +1
Query: 48 REKDANIKPDPDIDVYMKAVATEGQKENLITDYV 81
REK A IKPDPDID YMKA A Q+ +++TD V
Sbjct: 1069 REKHAKIKPDPDIDAYMKAAALGRQRTSVVTDLV 1170
Score = 42.0 bits (97), Expect(2) = 4e-12
Identities = 18/24 (75%), Positives = 18/24 (75%)
Frame = +3
Query: 24 HWRNDC*RNLGLLS*SPRSWTSL* 47
HWRNDC RN G LS* PR WT L*
Sbjct: 975 HWRNDCERNSGFLS*MPRGWTEL* 1046
>BE806563 similar to GP|11994752|db ABC transporter-like protein {Arabidopsis
thaliana}, partial (20%)
Length = 428
Score = 63.2 bits (152), Expect = 1e-10
Identities = 44/110 (40%), Positives = 69/110 (62%), Gaps = 3/110 (2%)
Frame = +1
Query: 65 KAVATEGQKENLITDYVLRVLGLEICADTVVGN--AMIRAISGGQKKRLTTG-EMLVGPT 121
K+++ E +KE+ + V+ LGL C ++ VG A+ R ISGG++KR++ G EMLV P+
Sbjct: 109 KSLSREEKKEH--AEMVIAELGLTRCRNSPVGGCMALFRGISGGERKRVSIGQEMLVNPS 282
Query: 122 KALFMDEISTGLDSSTTFQIVNSMRQYVHILKGTVVISLLQPPPETYNLF 171
LF+DE ++GLD STT Q++ S+ + TVV ++ QP Y +F
Sbjct: 283 -LLFVDEPTSGLD-STTAQLIVSVLHGLARAGRTVVATIHQPSSRLYRMF 426
>TC208020 similar to UP|Q76CU2 (Q76CU2) PDR-type ABC transporter 1, partial
(14%)
Length = 765
Score = 61.6 bits (148), Expect = 4e-10
Identities = 44/149 (29%), Positives = 79/149 (52%), Gaps = 7/149 (4%)
Frame = +2
Query: 76 LITDYVLRVLGLEICADTVVGNAMIRAISGGQKKRLTTGEMLVGPTKALFMDEISTGLDS 135
+ + V+ ++ L +++VG + +S Q+KRLT LV +FMDE ++GLD+
Sbjct: 158 MFIEEVMELVELNPLRNSLVGLPGVSGLSTEQRKRLTIAVELVANPSIIFMDEPTSGLDA 337
Query: 136 STTFQIVNSMRQYVHILKGTVVISLLQPPPETYNLFDDIILLS-DSHIIYQGP----REH 190
++ ++R V + TVV ++ QP + + FD++ L+ IY GP H
Sbjct: 338 RAAAIVMRTVRNTVDTGR-TVVCTIHQPSIDIFEAFDELFLMKRGGQEIYVGPLGRHSTH 514
Query: 191 VLEFFESIG--FKCPNRKGVADFLQEVTS 217
++++FESIG K + A ++ EVT+
Sbjct: 515 LIKYFESIGGVSKIKDGYNPATWMLEVTT 601
>BI892553
Length = 421
Score = 57.0 bits (136), Expect = 1e-08
Identities = 36/97 (37%), Positives = 51/97 (52%), Gaps = 5/97 (5%)
Frame = +3
Query: 132 GLDSSTTFQIVNSM-----RQYVHILKGTVVISLLQPPPETYNLFDDIILLSDSHIIYQG 186
GLDS+ ++ ++ + + VH TVV S+ QP E + LFD++ LLS +Y G
Sbjct: 3 GLDSAASYYVMKRIATLDKKDDVH---RTVVASIHQPSSEVFQLFDNLCLLSSGRTVYFG 173
Query: 187 PREHVLEFFESIGFKCPNRKGVADFLQEVTSRKDQEQ 223
P EFF S GF CP +D L + T KD +Q
Sbjct: 174 PASAAKEFFASNGFPCPPLMNPSDHLLK-TINKDFDQ 281
>TC207596 similar to UP|Q7GBY1 (Q7GBY1) Tonoplast ABC transporter IDI7,
partial (35%)
Length = 1076
Score = 47.4 bits (111), Expect = 8e-06
Identities = 28/100 (28%), Positives = 54/100 (54%)
Frame = +2
Query: 93 TVVGNAMIRAISGGQKKRLTTGEMLVGPTKALFMDEISTGLDSSTTFQIVNSMRQYVHIL 152
T VG +R +SGGQK+R+ L+ K L +DE ++ LD+ + + + ++M ++
Sbjct: 329 TFVGERGVR-LSGGQKQRIAIARALLMDPKILLLDEATSALDAESEYLVQDAMES---LM 496
Query: 153 KGTVVISLLQPPPETYNLFDDIILLSDSHIIYQGPREHVL 192
KG V+ ++ T D + ++SD ++ +G E +L
Sbjct: 497 KGRTVL-VIAHRLSTVKTADTVAVISDGQVVERGNHEELL 613
>TC218748 similar to UP|Q76CU1 (Q76CU1) PDR-type ABC transporter 2
(Fragment), partial (37%)
Length = 1384
Score = 45.4 bits (106), Expect = 3e-05
Identities = 36/118 (30%), Positives = 63/118 (52%), Gaps = 8/118 (6%)
Frame = +3
Query: 113 TGEMLVGPTKALFMDEISTGLDSSTTFQIVNSMRQYVHILKGTVVISLLQPPPETYNLFD 172
T E++ P+ +FMDE ++GLD+ ++ ++R V + TVV ++ QP + + FD
Sbjct: 3 TSELVANPS-IIFMDEPTSGLDARAAAIVMRTVRNTVDTGR-TVVCTIHQPSIDIFESFD 176
Query: 173 DIILLSD-SHIIYQGP----REHVLEFFESIG--FKCPNRKGVADFLQEV-TSRKDQE 222
+++L+ IY GP H++ +FE I K + A ++ EV TS K+ E
Sbjct: 177 ELLLMKQGGQEIYVGPLGHHSSHLINYFEGIQGVNKIKDGYNPATWMLEVSTSAKEME 350
>TC219347 similar to UP|Q9LF78 (Q9LF78) Mitochondrial half-ABC transporter,
partial (36%)
Length = 1085
Score = 44.7 bits (104), Expect = 5e-05
Identities = 29/100 (29%), Positives = 52/100 (52%)
Frame = +1
Query: 93 TVVGNAMIRAISGGQKKRLTTGEMLVGPTKALFMDEISTGLDSSTTFQIVNSMRQYVHIL 152
TVVG ++ +SGG+K+R+ + L DE ++ LDS+T +I+++++ +
Sbjct: 436 TVVGERGLK-LSGGEKQRVALARAFLKAPAILLCDEATSALDSTTEAEILSALKSVANNR 612
Query: 153 KGTVVISLLQPPPETYNLFDDIILLSDSHIIYQGPREHVL 192
+ L T D+II+L + +I QGP E +L
Sbjct: 613 TSIFIAHRL----TTAMQCDEIIVLENGKVIEQGPHEVLL 720
>TC218050 weakly similar to UP|Q7PC82 (Q7PC82) PDR14 ABC transporter, partial
(28%)
Length = 1436
Score = 43.5 bits (101), Expect = 1e-04
Identities = 27/87 (31%), Positives = 49/87 (56%), Gaps = 5/87 (5%)
Frame = +2
Query: 117 LVGPTKALFMDEISTGLDSSTTFQIVNSMRQYVHILKGTVVISLLQPPPETYNLFDDIIL 176
LV +FMDE ++GLD+ ++ +++ V + T V ++ QP + + FD++IL
Sbjct: 14 LVSNPSIIFMDEPTSGLDARAAAVVMRAVKNVVATGR-TTVCTIHQPSIDIFETFDELIL 190
Query: 177 L-SDSHIIYQGPREH----VLEFFESI 198
+ S IIY G H ++E+F++I
Sbjct: 191 MKSGGRIIYSGMLGHHSSRLIEYFQNI 271
>BI699796 similar to PIR|G85167|G85 ABC transporter like protein [imported] -
Arabidopsis thaliana, partial (13%)
Length = 421
Score = 42.7 bits (99), Expect = 2e-04
Identities = 23/69 (33%), Positives = 38/69 (54%)
Frame = +1
Query: 81 VLRVLGLEICADTVVGNAMIRAISGGQKKRLTTGEMLVGPTKALFMDEISTGLDSSTTFQ 140
V+ + L+ D++VG I +S Q+KRLT LV +FMDE +TGLD+
Sbjct: 187 VIHTIELDGIKDSLVGMPNISGLSTEQRKRLTIAVELVANPSIIFMDEPTTGLDARAAAV 366
Query: 141 IVNSMRQYV 149
++ +++ V
Sbjct: 367 VMRAVKNVV 393
>TC212091 similar to UP|Q9LFG8 (Q9LFG8) ABC transporter-like protein, partial
(12%)
Length = 477
Score = 42.0 bits (97), Expect = 3e-04
Identities = 24/93 (25%), Positives = 50/93 (52%)
Frame = +3
Query: 94 VVGNAMIRAISGGQKKRLTTGEMLVGPTKALFMDEISTGLDSSTTFQIVNSMRQYVHILK 153
V+G+ R++SGG+++R++ G ++ LF+DE ++ LDS++ F +V +++
Sbjct: 9 VIGDNGHRSVSGGERRRVSIGTDIIHNPIVLFLDEPTSNLDSTSAFMVVKVLQRIAQ--S 182
Query: 154 GTVVISLLQPPPETYNLFDDIILLSDSHIIYQG 186
G + I + P +L + L H + +G
Sbjct: 183 GNIFIMSIH*PSYRISLRPLDLPLPRQHRLQRG 281
>TC207459 similar to UP|Q94IH6 (Q94IH6) CjMDR1, partial (30%)
Length = 1329
Score = 40.0 bits (92), Expect = 0.001
Identities = 29/109 (26%), Positives = 56/109 (50%)
Frame = +1
Query: 84 VLGLEICADTVVGNAMIRAISGGQKKRLTTGEMLVGPTKALFMDEISTGLDSSTTFQIVN 143
+ GL+ DT+VG + +SGGQK+R+ ++ K L +DE ++ LD+ + ++V
Sbjct: 784 ISGLQQGYDTIVGERGTQ-LSGGQKQRVAIARAIIKSPKILLLDEATSALDAESE-RVVQ 957
Query: 144 SMRQYVHILKGTVVISLLQPPPETYNLFDDIILLSDSHIIYQGPREHVL 192
V + + TVV++ T D I ++ + I+ +G E ++
Sbjct: 958 DALDKVMVNRTTVVVA---HRLSTIKNADVIAVVKNGVIVEKGKHEKLI 1095
>TC232373 weakly similar to UP|AB10_MOUSE (Q9JI39) ATP-binding cassette,
sub-family B, member 10, mitochondrial precursor
(ATP-binding cassette transporter 10) (ABC transporter
10 protein) (ABC-mitochondrial erythroid protein)
(ABC-me protein), partial (24%)
Length = 742
Score = 38.5 bits (88), Expect = 0.004
Identities = 29/101 (28%), Positives = 51/101 (49%)
Frame = +1
Query: 92 DTVVGNAMIRAISGGQKKRLTTGEMLVGPTKALFMDEISTGLDSSTTFQIVNSMRQYVHI 151
DT+VG I+ +SGGQK+R+ +V K L +DE ++ LD + ++V V +
Sbjct: 322 DTIVGERGIQ-LSGGQKQRVAIARAIVKNPKILLLDEATSALDVESE-RVVQDALDQVMV 495
Query: 152 LKGTVVISLLQPPPETYNLFDDIILLSDSHIIYQGPREHVL 192
+ T+V++ T D I ++ + I QG + +L
Sbjct: 496 DRTTIVVA---HRLSTIKDADSIAVVQNGVIAEQGKHDTLL 609
>TC207756 similar to UP|Q8RY46 (Q8RY46) Transporter associated with antigen
processing-like protein, partial (17%)
Length = 785
Score = 37.0 bits (84), Expect = 0.011
Identities = 22/90 (24%), Positives = 41/90 (45%)
Frame = +3
Query: 103 ISGGQKKRLTTGEMLVGPTKALFMDEISTGLDSSTTFQIVNSMRQYVHILKGTVVISLLQ 162
+SGGQK+R+ L+ K L +DE ++ LD+ + + +R V T + ++
Sbjct: 45 LSGGQKQRIAIARALLRDPKILILDEATSALDAESEHNVKGVLRS-VRSDSATRSVIVIA 221
Query: 163 PPPETYNLFDDIILLSDSHIIYQGPREHVL 192
T D I+++ I+ G +L
Sbjct: 222 HRLSTIQAADRIVVMDGGEIVEMGSHRELL 311
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.327 0.141 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,733,689
Number of Sequences: 63676
Number of extensions: 162203
Number of successful extensions: 963
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 949
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 956
length of query: 296
length of database: 12,639,632
effective HSP length: 97
effective length of query: 199
effective length of database: 6,463,060
effective search space: 1286148940
effective search space used: 1286148940
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148607.1


