

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148532.8 + phase: 0
(380 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
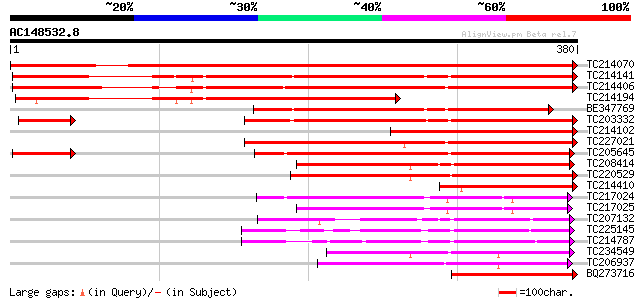
Score E
Sequences producing significant alignments: (bits) Value
TC214070 similar to UP|Q8VWQ1 (Q8VWQ1) Dehydration-induced prote... 541 e-154
TC214141 similar to UP|Q947D4 (Q947D4) Dehydration-responsive pr... 476 e-134
TC214406 weakly similar to UP|Q947D4 (Q947D4) Dehydration-respon... 470 e-133
TC214194 weakly similar to UP|Q94IC5 (Q94IC5) BURP domain-contai... 266 1e-71
BE347769 weakly similar to GP|16588826|gb dehydration-responsive... 224 5e-59
TC203332 weakly similar to UP|Q8H9E9 (Q8H9E9) Resistant specific... 215 3e-56
TC214102 similar to UP|Q9MB24 (Q9MB24) S1-3 protein (Fragment), ... 208 3e-54
TC227021 weakly similar to UP|O65009 (O65009) BURP domain contai... 206 2e-53
TC205645 UP|Q8RUD9 (Q8RUD9) Seed coat BURP domain protein 1, com... 189 1e-48
TC208414 weakly similar to UP|O65009 (O65009) BURP domain contai... 156 1e-38
TC220529 weakly similar to GB|CAE02618.1|38567637|OSA575668 RAFT... 152 3e-37
TC214410 similar to UP|Q8VWQ1 (Q8VWQ1) Dehydration-induced prote... 147 8e-36
TC217024 similar to UP|Q40161 (Q40161) Polygalacturonase isoenzy... 142 2e-34
TC217025 similar to UP|Q40161 (Q40161) Polygalacturonase isoenzy... 138 5e-33
TC207132 similar to UP|EA92_VICFA (P21747) Embryonic abundant pr... 135 3e-32
TC225145 homologue to UP|O24482 (O24482) Sali3-2, complete 129 2e-30
TC214787 UP|Q06765 (Q06765) ADR6 protein (Sali5-4a protein), com... 124 9e-29
TC234549 weakly similar to UP|O65009 (O65009) BURP domain contai... 119 3e-27
TC206937 similar to UP|Q40161 (Q40161) Polygalacturonase isoenzy... 117 1e-26
BQ273716 weakly similar to SP|Q08298|RD22_ Dehydration-responsiv... 113 1e-25
>TC214070 similar to UP|Q8VWQ1 (Q8VWQ1) Dehydration-induced protein RD22-like
protein, partial (81%)
Length = 1427
Score = 541 bits (1394), Expect = e-154
Identities = 274/383 (71%), Positives = 312/383 (80%), Gaps = 3/383 (0%)
Frame = +3
Query: 1 MCVYVVLQLALVATHA-ALPPEVYWKSKLPTTQMPKAITDLLHLDWTEEKSTSVAVGKGG 59
+ ++ +L LA+VATHA LPPEVYWKSKLPTT MPKAITD+LH D E+KSTSVAVGKG
Sbjct: 111 LSIFALLNLAVVATHAETLPPEVYWKSKLPTTPMPKAITDILHPDLAEDKSTSVAVGKG- 287
Query: 60 VNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRVNV 119
GV+V+AGKTKPGGT+VNVGKGGV+VN GKGKP GT VNV
Sbjct: 288 --------------------GVNVNAGKTKPGGTSVNVGKGGVNVNTGKGKPNKGTSVNV 407
Query: 120 GKGGVNVHAGKGRGKPVHVSVGNKSPFLYNYAASETQLHDKPNVALFFLEKDLHQGTKLN 179
GKGGVNV+ G +GKPVHV VG SPF YNYAASETQ HD PNVALFFLEKDLH GTKLN
Sbjct: 408 GKGGVNVNTGPKKGKPVHVGVGPHSPFDYNYAASETQWHDDPNVALFFLEKDLHYGTKLN 587
Query: 180 LQFTKT-NSNDDAKFLPKEVANSIPFSSSKVENILNLFSIKEGTEESEIVKNTISECEEN 238
L FT+ S+ DA FLP+ VA+SIPFSS+KV +LN FSIKEG++E++ VKNTISECE
Sbjct: 588 LHFTRYFTSSVDASFLPRSVADSIPFSSNKVNEVLNKFSIKEGSDEAQTVKNTISECEVP 767
Query: 239 GIKGEEKLCVTSLESMVDFTTSKLGNN-VEAVSTEVKKESSDLQEYVMAKGVKKLGEKNK 297
GIKGEEK CVTSLESMVDF T+KLG+N V+AVSTEV K+ ++LQ+Y MA GVK+LGE
Sbjct: 768 GIKGEEKRCVTSLESMVDFATTKLGSNDVDAVSTEVTKKDNELQQYTMAPGVKRLGEDKA 947
Query: 298 AVVCHKESYPYAVFYCHKTDSTKVYSVPLEGVDGSRVKAVAVCHTDTSQWNPKHLAFQVL 357
+VVCHKE+YPYAVFYCHK+++TK YSVPLEG DGSRVKAVAVCHTDTS+WNPKHLAFQVL
Sbjct: 948 SVVCHKENYPYAVFYCHKSETTKAYSVPLEGADGSRVKAVAVCHTDTSKWNPKHLAFQVL 1127
Query: 358 NVQPGTVPVCHFLPQDHVVWVSK 380
VQPGTVPVCHFLPQDHVV+V K
Sbjct: 1128KVQPGTVPVCHFLPQDHVVFVPK 1196
>TC214141 similar to UP|Q947D4 (Q947D4) Dehydration-responsive protein RD22
(Fragment), partial (89%)
Length = 1353
Score = 476 bits (1224), Expect = e-134
Identities = 249/381 (65%), Positives = 290/381 (75%), Gaps = 3/381 (0%)
Frame = +1
Query: 3 VYVVLQLALVATHAALPPEVYWKSKLPTTQMPKAITDLLHLDWTEEKSTSVAVGKGGVNV 62
++ +L +ALVATHAALPPEVYWKS LPTT MPKAITD+L+ DW EEKS+SV
Sbjct: 97 IFTLLNIALVATHAALPPEVYWKSVLPTTPMPKAITDILYSDWVEEKSSSV--------- 249
Query: 63 DSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRVNVGK- 121
+VG GGV+V+ GKG G GT VNVG
Sbjct: 250 ---------------------------------HVGGGGVNVHTGKGG-GSGTTVNVGGK 327
Query: 122 --GGVNVHAGKGRGKPVHVSVGNKSPFLYNYAASETQLHDKPNVALFFLEKDLHQGTKLN 179
GGVNVHAG +GKPVHVSVG+KSPF Y YAA+ETQLHD PNVALFFLEKDLH GTKL+
Sbjct: 328 GGGGVNVHAGH-KGKPVHVSVGSKSPFDYVYAATETQLHDDPNVALFFLEKDLHSGTKLD 504
Query: 180 LQFTKTNSNDDAKFLPKEVANSIPFSSSKVENILNLFSIKEGTEESEIVKNTISECEENG 239
L FT++ SN A FL ++VA+SIPFSS+KV+ I N FS+K G+EE++I+KNTISECEE G
Sbjct: 505 LHFTRSTSNQ-ATFLSRQVADSIPFSSNKVDFIFNKFSVKPGSEEAQIMKNTISECEEGG 681
Query: 240 IKGEEKLCVTSLESMVDFTTSKLGNNVEAVSTEVKKESSDLQEYVMAKGVKKLGEKNKAV 299
IKGEEK C TSLESMVDF+TSKLGNNVE VSTEV KE+ LQ+Y +A GVKKL +KAV
Sbjct: 682 IKGEEKYCATSLESMVDFSTSKLGNNVEVVSTEVDKETG-LQKYTVAPGVKKLSG-DKAV 855
Query: 300 VCHKESYPYAVFYCHKTDSTKVYSVPLEGVDGSRVKAVAVCHTDTSQWNPKHLAFQVLNV 359
VCHK++YPYAVFYCHKT++T+ YSVPLEG +G RVKAVAVCHTDTS+WNPKHLAFQVL V
Sbjct: 856 VCHKQNYPYAVFYCHKTETTRAYSVPLEGTNGVRVKAVAVCHTDTSEWNPKHLAFQVLKV 1035
Query: 360 QPGTVPVCHFLPQDHVVWVSK 380
+PGT+PVCHFLP+DHVVWV K
Sbjct: 1036KPGTIPVCHFLPEDHVVWVPK 1098
>TC214406 weakly similar to UP|Q947D4 (Q947D4) Dehydration-responsive protein
RD22 (Fragment), partial (90%)
Length = 1379
Score = 470 bits (1210), Expect = e-133
Identities = 248/383 (64%), Positives = 286/383 (73%), Gaps = 5/383 (1%)
Frame = +1
Query: 3 VYVVLQLALVATHAALPPEVYWKSKLPTTQMPKAITDLLHLDWTEEKSTSVAVGKGGVNV 62
++ +L LALVA HAALPPEVYWKS LPTT MPKAITD+L+ DW EEKSTSV VG GVNV
Sbjct: 145 IFTLLNLALVAIHAALPPEVYWKSVLPTTPMPKAITDILYPDWVEEKSTSVNVGGKGVNV 324
Query: 63 DSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRVNVG-- 120
+ GKG GGGT VNVG
Sbjct: 325 ---------------------------------HAGKG-----------GGGTNVNVGGK 372
Query: 121 --KGGVNVHAGKGRGKPVHVSVGNKSPFLYNYAASETQLHDKPNVALFFLEKDLHQGTKL 178
GGVNVHAG +GKPVHVSVG+KSPF Y YA++ETQLHD PNVALFFLEKDLH GTKL
Sbjct: 373 GSGGGVNVHAGH-KGKPVHVSVGSKSPFNYIYASTETQLHDDPNVALFFLEKDLHPGTKL 549
Query: 179 NLQFTKTNSNDDAKFLPKEVANSIPFSSSKVENILNLFSIKEGTEESEIVKNTISECEEN 238
NL FT T+SN A FLP++VA+SIPFSSSKVE + N FS+K G+EE++I+KNT+SECEE
Sbjct: 550 NLHFT-TSSNIQATFLPRQVADSIPFSSSKVEVVFNKFSVKPGSEEAQIMKNTLSECEEG 726
Query: 239 GIKGEEKLCVTSLESMVDFTTSKLGNNVEAVSTE-VKKESSDLQEYVMAKGVKKLGEKNK 297
GIKGEEK C TSLESM+DF+TSKLG NVE VSTE V+ + + LQ+Y +A GV KL +K
Sbjct: 727 GIKGEEKYCATSLESMIDFSTSKLGKNVEVVSTEVVEDKETGLQKYTVAPGVNKL-SGDK 903
Query: 298 AVVCHKESYPYAVFYCHKTDSTKVYSVPLEGVDGSRVKAVAVCHTDTSQWNPKHLAFQVL 357
AVVCHK++YPYAVFYCHKT++T+ YSVPLEG +G RVKAVAVCHT TS+WNPKHLAFQVL
Sbjct: 904 AVVCHKQNYPYAVFYCHKTETTRAYSVPLEGANGVRVKAVAVCHTHTSEWNPKHLAFQVL 1083
Query: 358 NVQPGTVPVCHFLPQDHVVWVSK 380
V+PGTVPVCHFLP+DHVVWV K
Sbjct: 1084KVKPGTVPVCHFLPEDHVVWVPK 1152
>TC214194 weakly similar to UP|Q94IC5 (Q94IC5) BURP domain-containing protein
(Fragment), partial (49%)
Length = 704
Score = 266 bits (680), Expect = 1e-71
Identities = 154/265 (58%), Positives = 174/265 (65%), Gaps = 7/265 (2%)
Frame = +1
Query: 5 VVLQLALVATHA--ALPPEVYWKSKLPTTQMPKAITDLLHLDWTEEKSTSVAVGKGGVNV 62
+ + +VAT+A ALPP YWKSKLPTT MPKAITDLL DW EEK TSV
Sbjct: 37 ITFLMLVVATNADAALPPAFYWKSKLPTTPMPKAITDLLQPDWKEEKDTSV--------- 189
Query: 63 DSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGK--PGGGTRVNVG 120
+VGKGGV+V KG PG GT VNVG
Sbjct: 190 ---------------------------------DVGKGGVNVGVEKGNQGPGDGTDVNVG 270
Query: 121 --KGGVNVHAGKGRGKPVHVSVGNKSPFLYNYAASETQLHDKPNVALFFLEKDLHQGTKL 178
GGVNVH G +GKPVHV VG SPF YNYAASETQLHD PNVALFFLEKDLH GTKL
Sbjct: 271 GGDGGVNVHTGP-KGKPVHVGVGPHSPFDYNYAASETQLHDDPNVALFFLEKDLHHGTKL 447
Query: 179 NLQFT-KTNSNDDAKFLPKEVANSIPFSSSKVENILNLFSIKEGTEESEIVKNTISECEE 237
NL FT SN DA FLP+ V++SIPFSS+KV ++LN FSIK+G++E++ VKNTI+ECE
Sbjct: 448 NLHFTIYYTSNVDATFLPRSVSDSIPFSSNKVNDVLNKFSIKDGSDEAKTVKNTINECEG 627
Query: 238 NGIKGEEKLCVTSLESMVDFTTSKL 262
IKGEEK CVTSLESMVDF T+KL
Sbjct: 628 PSIKGEEKRCVTSLESMVDFATTKL 702
>BE347769 weakly similar to GP|16588826|gb dehydration-responsive protein
RD22 {Prunus persica}, partial (38%)
Length = 595
Score = 224 bits (571), Expect = 5e-59
Identities = 120/201 (59%), Positives = 151/201 (74%)
Frame = +2
Query: 164 ALFFLEKDLHQGTKLNLQFTKTNSNDDAKFLPKEVANSIPFSSSKVENILNLFSIKEGTE 223
AL+ +E DLH GTKL+L FT++ SN A FL ++VA+SIPFSS+KV+ I N S K G++
Sbjct: 2 ALYLVEFDLHSGTKLDLHFTRSTSNQ-ATFLSRQVADSIPFSSNKVDFIFNKNSGKPGSK 178
Query: 224 ESEIVKNTISECEENGIKGEEKLCVTSLESMVDFTTSKLGNNVEAVSTEVKKESSDLQEY 283
E++I+KNTI ECEE GIKGEEK C TSLESM DF+TSKLGNNVE VSTEV K++ +Y
Sbjct: 179 EAQIMKNTIIECEEGGIKGEEKYCATSLESMDDFSTSKLGNNVEVVSTEVDKDTG-*HKY 355
Query: 284 VMAKGVKKLGEKNKAVVCHKESYPYAVFYCHKTDSTKVYSVPLEGVDGSRVKAVAVCHTD 343
A GVKK+ +KAVVCHK +YPYA FYC++TD+T YSVP EG + R KAV+ CHTD
Sbjct: 356 TGAPGVKKV-SGDKAVVCHKHNYPYADFYCNRTDTTIAYSVPSEGTNVVRDKAVSRCHTD 532
Query: 344 TSQWNPKHLAFQVLNVQPGTV 364
TS P+H+A VL V P ++
Sbjct: 533 TSDMYPRHVANHVLTVIPRSI 595
>TC203332 weakly similar to UP|Q8H9E9 (Q8H9E9) Resistant specific protein-2,
partial (60%)
Length = 1444
Score = 215 bits (547), Expect = 3e-56
Identities = 104/223 (46%), Positives = 141/223 (62%)
Frame = +1
Query: 158 HDKPNVALFFLEKDLHQGTKLNLQFTKTNSNDDAKFLPKEVANSIPFSSSKVENILNLFS 217
HD P F E+ L GTKL+ F K + LP+++A IP SS+K++ I+ +
Sbjct: 583 HDIPKADQVFFEEGLRPGTKLDAHFKKRENV--TPLLPRQIAQHIPLSSAKIKEIVEMLF 756
Query: 218 IKEGTEESEIVKNTISECEENGIKGEEKLCVTSLESMVDFTTSKLGNNVEAVSTEVKKES 277
+ E +I++ TIS CE I GEE+ C TSLESMVDF TSKLG N +STE +KES
Sbjct: 757 VNPEPENVKILEETISMCEVPAITGEERYCATSLESMVDFVTSKLGKNARVISTEAEKES 936
Query: 278 SDLQEYVMAKGVKKLGEKNKAVVCHKESYPYAVFYCHKTDSTKVYSVPLEGVDGSRVKAV 337
Q++ + GVK L E +K +VCH YPY VF CH+ +T + +PLEG DG+RVKA
Sbjct: 937 KS-QKFSVKDGVKLLAE-DKVIVCHPMDYPYVVFMCHEISNTTAHFMPLEGEDGTRVKAA 1110
Query: 338 AVCHTDTSQWNPKHLAFQVLNVQPGTVPVCHFLPQDHVVWVSK 380
AVCH DTS+W+P H+ Q+L +PG PVCH P+ H++W +K
Sbjct: 1111AVCHKDTSEWDPNHVFLQMLKTKPGAAPVCHIFPEGHLLWFAK 1239
Score = 54.7 bits (130), Expect = 7e-08
Identities = 24/38 (63%), Positives = 28/38 (73%)
Frame = +1
Query: 7 LQLALVATHAALPPEVYWKSKLPTTQMPKAITDLLHLD 44
L L L+ HAA+PPEVYW+ LP T MPKAI D L+LD
Sbjct: 46 LNLLLMTAHAAIPPEVYWERMLPNTPMPKAIIDFLNLD 159
>TC214102 similar to UP|Q9MB24 (Q9MB24) S1-3 protein (Fragment), partial
(70%)
Length = 621
Score = 208 bits (530), Expect = 3e-54
Identities = 96/126 (76%), Positives = 112/126 (88%), Gaps = 1/126 (0%)
Frame = +3
Query: 256 DFTTSKLGNN-VEAVSTEVKKESSDLQEYVMAKGVKKLGEKNKAVVCHKESYPYAVFYCH 314
DF T+KLG+N V+AVSTEV K+ ++LQ+Y MA GVK+LGE +VVCHKE+YPYAVFYCH
Sbjct: 15 DFATTKLGSNDVDAVSTEVTKKDNELQQYTMAPGVKRLGEDKASVVCHKENYPYAVFYCH 194
Query: 315 KTDSTKVYSVPLEGVDGSRVKAVAVCHTDTSQWNPKHLAFQVLNVQPGTVPVCHFLPQDH 374
K+++TK YSVPLEG DGSRVKAVAVCHTDTS+WNPKHLAFQVL V PGTVP+CHFLPQDH
Sbjct: 195 KSETTKAYSVPLEGADGSRVKAVAVCHTDTSKWNPKHLAFQVLKVHPGTVPICHFLPQDH 374
Query: 375 VVWVSK 380
VV+V K
Sbjct: 375 VVFVPK 392
>TC227021 weakly similar to UP|O65009 (O65009) BURP domain containing
protein, partial (47%)
Length = 1147
Score = 206 bits (523), Expect = 2e-53
Identities = 102/227 (44%), Positives = 147/227 (63%), Gaps = 4/227 (1%)
Frame = +1
Query: 158 HDKPNVALFFLEKDLHQGTKLNLQFTKTNSNDDAKFLPKEVANSIPFSSSKVENILNLFS 217
H P+V +FF +DL G + + F K + K P+E A+S+PFS +K+ N+L +FS
Sbjct: 295 HIDPSVMVFFTIEDLKVGKTMPIHFPKRDPATSPKLWPREEADSLPFSLNKLPNLLKIFS 474
Query: 218 IKEGTEESEIVKNTISECEENGIKGEEKLCVTSLESMVDFTTSKLG--NNVEAVSTEVKK 275
+ + + +++ +++T+ ECE IKGE K C TSLESM+DFT S LG ++++ +ST +
Sbjct: 475 VSQNSPKAKAMEDTLRECETKPIKGEVKFCATSLESMLDFTQSILGFTSDLKVLSTSHQT 654
Query: 276 ESS-DLQEYVMAKGVKKLGEKNKAVVCHKESYPYAVFYCHKTDS-TKVYSVPLEGVDGSR 333
+SS Q Y M + + ++ +K V CH YPY VFYCH +S K+Y VPL G +G R
Sbjct: 655 KSSVTFQNYTMLENIIEI-PASKMVACHTMPYPYTVFYCHSQESENKIYRVPLAGENGDR 831
Query: 334 VKAVAVCHTDTSQWNPKHLAFQVLNVQPGTVPVCHFLPQDHVVWVSK 380
V A+ VCH DTSQW H++FQVL V+PGT VCHF P DH++WV K
Sbjct: 832 VDAMVVCHMDTSQWGHGHVSFQVLKVKPGTTSVCHFFPADHLIWVPK 972
>TC205645 UP|Q8RUD9 (Q8RUD9) Seed coat BURP domain protein 1, complete
Length = 1369
Score = 189 bits (481), Expect = 1e-48
Identities = 93/214 (43%), Positives = 133/214 (61%)
Frame = +1
Query: 165 LFFLEKDLHQGTKLNLQFTKTNSNDDAKFLPKEVANSIPFSSSKVENILNLFSIKEGTEE 224
LFFLE+DL G N++F N+ LP++++ IPFS K + +L + ++ +
Sbjct: 310 LFFLEEDLRAGKIFNMKFVN-NTKATVPLLPRQISKQIPFSEDKKKQVLAMLGVEANSSN 486
Query: 225 SEIVKNTISECEENGIKGEEKLCVTSLESMVDFTTSKLGNNVEAVSTEVKKESSDLQEYV 284
++I+ TI C+E +GE K C TSLESMVDF S LG NV A STE ++E+ + V
Sbjct: 487 AKIIAETIGLCQEPATEGERKHCATSLESMVDFVVSALGKNVGAFSTEKERETESGKFVV 666
Query: 285 MAKGVKKLGEKNKAVVCHKESYPYAVFYCHKTDSTKVYSVPLEGVDGSRVKAVAVCHTDT 344
+ GV+KLG+ +K + CH SYPY VF CH + Y V L+G DG RVKAV CH DT
Sbjct: 667 VKNGVRKLGD-DKVIACHPMSYPYVVFGCHLVPRSSGYLVRLKGEDGVRVKAVVACHRDT 843
Query: 345 SQWNPKHLAFQVLNVQPGTVPVCHFLPQDHVVWV 378
S+W+ H AF+VLN++PG VCH + +++W+
Sbjct: 844 SKWDHNHGAFKVLNLKPGNGTVCHVFTEGNLLWL 945
Score = 46.6 bits (109), Expect = 2e-05
Identities = 20/42 (47%), Positives = 30/42 (70%)
Frame = +1
Query: 3 VYVVLQLALVATHAALPPEVYWKSKLPTTQMPKAITDLLHLD 44
+++ L L L+ +AAL P YW++ LP T +PKAIT+LL L+
Sbjct: 58 IFLYLNLMLMTANAALTPRHYWETMLPRTPLPKAITELLSLE 183
>TC208414 weakly similar to UP|O65009 (O65009) BURP domain containing
protein, partial (39%)
Length = 957
Score = 156 bits (395), Expect = 1e-38
Identities = 79/190 (41%), Positives = 118/190 (61%), Gaps = 4/190 (2%)
Frame = +2
Query: 193 FLPKEVANSIPFSSSKVENILNLFSIKEGTEESEIVKNTISECEENGIKGEEKLCVTSLE 252
F P++ FSS + ++L FSI + + +++ +K T+ +CE ++GE K C TSLE
Sbjct: 239 F*PEKKLTRFLFSSKHLPSLLKFFSIPKHSPQAKAMKYTLKQCEFEPMEGETKFCATSLE 418
Query: 253 SMVDFTTSKLGNNVE---AVSTEVKKESSDLQEYVMAKGVKKLGEKNKAVVCHKESYPYA 309
S+ DF G+N + + + ++ LQ Y +++ VK + N + CH YPYA
Sbjct: 419 SLFDFAHYLFGSNAQFKVLTTVHLTNSTTLLQNYTISE-VKVISVPN-VIGCHPMPYPYA 592
Query: 310 VFYCHKTDS-TKVYSVPLEGVDGSRVKAVAVCHTDTSQWNPKHLAFQVLNVQPGTVPVCH 368
VFYCH S T +Y V +EG +G RV+A A+CH DTS+W+ H++F+VL VQPGT PVCH
Sbjct: 593 VFYCHSQHSDTNLYEVMVEGENGGRVQAAAICHMDTSKWDRDHVSFRVLKVQPGTSPVCH 772
Query: 369 FLPQDHVVWV 378
F P D++VWV
Sbjct: 773 FFPPDNLVWV 802
Score = 38.1 bits (87), Expect = 0.007
Identities = 25/84 (29%), Positives = 37/84 (43%)
Frame = +1
Query: 121 KGGVNVHAGKGRGKPVHVSVGNKSPFLYNYAASETQLHDKPNVALFFLEKDLHQGTKLNL 180
K G+ + K H + K Y + E H + +FF DL G + +
Sbjct: 25 KKGIGFDIEEEEDKNDHKKIVGKDVHGYK-PSHEDHKHMDLELNVFFTPNDLKVGKIMPI 201
Query: 181 QFTKTNSNDDAKFLPKEVANSIPF 204
F+K NS+ KFL +E A+ IPF
Sbjct: 202 YFSKKNSSTSPKFLTREEADQIPF 273
>TC220529 weakly similar to GB|CAE02618.1|38567637|OSA575668 RAFTIN1 protein
{Oryza sativa (japonica cultivar-group);} , partial
(15%)
Length = 942
Score = 152 bits (383), Expect = 3e-37
Identities = 75/197 (38%), Positives = 121/197 (61%), Gaps = 5/197 (2%)
Frame = +3
Query: 189 DDAKFLPKEVANSIPFSSSKVENILNLFSIKEGTEESEIVKNTISECEENGIKGEEKLCV 248
D ++FLP++ A SIPFS S++ ++L LFSI E + E+ +++T+ +CE I GE K+C
Sbjct: 204 DVSQFLPRKEAESIPFSISQLPSVLQLFSISEDSPEANAMRDTLEQCEAEPITGETKICA 383
Query: 249 TSLESMVDFTTSKLGNNVE---AVSTEVKKESSDLQEYVMAKGVKKLGEKNKAVVCHKES 305
TSLESM++F + +G+ + +T LQ++ + + + + K V CH
Sbjct: 384 TSLESMLEFIGTIIGSETKHNILTTTLPTASGVPLQKFTILEVSEDINAA-KWVACHPLP 560
Query: 306 YPYAVFYCH-KTDSTKVYSVPLEGVDG-SRVKAVAVCHTDTSQWNPKHLAFQVLNVQPGT 363
YPYA++YCH +KV+ V L +G +++A+ +CH DTS W+P H+ F+ L ++PG
Sbjct: 561 YPYAIYYCHFIATGSKVFKVSLGSENGDDKIEALGICHLDTSDWSPNHIIFRQLGIKPGK 740
Query: 364 VPVCHFLPQDHVVWVSK 380
VCHF P H++WV K
Sbjct: 741 DAVCHFFPIKHLMWVPK 791
>TC214410 similar to UP|Q8VWQ1 (Q8VWQ1) Dehydration-induced protein RD22-like
protein, partial (22%)
Length = 575
Score = 147 bits (371), Expect = 8e-36
Identities = 70/96 (72%), Positives = 79/96 (81%), Gaps = 4/96 (4%)
Frame = +1
Query: 289 VKKLGEKNKAVVC----HKESYPYAVFYCHKTDSTKVYSVPLEGVDGSRVKAVAVCHTDT 344
+K L +KN +V + PYAVFYCHK+++TK YSVPLEG DGSRVKAVAVCHTDT
Sbjct: 55 IKILQKKNSRLVLSLVPNSARGPYAVFYCHKSETTKAYSVPLEGADGSRVKAVAVCHTDT 234
Query: 345 SQWNPKHLAFQVLNVQPGTVPVCHFLPQDHVVWVSK 380
S+WNPKHLAFQVL VQPGTVPVCHFLPQDHVV+V K
Sbjct: 235 SKWNPKHLAFQVLKVQPGTVPVCHFLPQDHVVFVPK 342
>TC217024 similar to UP|Q40161 (Q40161) Polygalacturonase isoenzyme 1 beta
subunit precursor , partial (33%)
Length = 958
Score = 142 bits (359), Expect = 2e-34
Identities = 77/217 (35%), Positives = 122/217 (55%), Gaps = 5/217 (2%)
Frame = +1
Query: 166 FFLEKDLHQGTKLNLQFTKTNSNDDAKFLPKEVANSIPFSSSKVENILNLFSIKEGTEES 225
FF E L +GT + + + + FLP+ + +PFSS+KV+ + +F + E +
Sbjct: 85 FFRETMLKEGTVMPMPDIR-DKMPKRSFLPRAILTKLPFSSAKVDELKRVFKVSENSSMD 261
Query: 226 EIVKNTISECEENGIKGEEKLCVTSLESMVDFTTSKLGNNVEAVSTEVKKESSDLQEYVM 285
+++ +++ ECE GE K CV S+E M+DF+TS LG NV +TE K S+ + VM
Sbjct: 262 KMIMDSLGECERAPSVGETKRCVASVEDMIDFSTSVLGRNVAVWTTENVKGSN---KNVM 432
Query: 286 AKGVKKL--GEKNKAVVCHKESYPYAVFYCHKTDSTKVYSVPLEGVDGSRVK---AVAVC 340
VK + G+ K+V CH+ +PY ++YCH +VY L + S+ K VA+C
Sbjct: 433 VGRVKGMNGGKVTKSVSCHQSLFPYLLYYCHSLPKVRVYEANLLDPE-SKAKINHGVAIC 609
Query: 341 HTDTSQWNPKHLAFQVLNVQPGTVPVCHFLPQDHVVW 377
H DT+ W+P H AF L PG + VCH++ ++ + W
Sbjct: 610 HLDTTAWSPTHGAFLALGSGPGRIEVCHWIFENDLTW 720
>TC217025 similar to UP|Q40161 (Q40161) Polygalacturonase isoenzyme 1 beta
subunit precursor , partial (32%)
Length = 933
Score = 138 bits (347), Expect = 5e-33
Identities = 70/190 (36%), Positives = 109/190 (56%), Gaps = 5/190 (2%)
Frame = +2
Query: 193 FLPKEVANSIPFSSSKVENILNLFSIKEGTEESEIVKNTISECEENGIKGEEKLCVTSLE 252
FLP+ + +PFSSSKV + LF + + + +++ +++ ECE GE K CV S+E
Sbjct: 44 FLPRSILTKLPFSSSKVHELKRLFKVSDNSSMEKMIMDSLGECERVPSMGETKRCVGSIE 223
Query: 253 SMVDFTTSKLGNNVEAVSTEVKKESSDLQEYVMAKGVKKL--GEKNKAVVCHKESYPYAV 310
M+DF+TS LG NV +TE S+ + VM VK + G+ ++V CH+ +PY +
Sbjct: 224 DMIDFSTSVLGRNVAVWTTENVNGSN---KNVMVGRVKGMNGGKVTQSVSCHQSLFPYML 394
Query: 311 FYCHKTDSTKVYSVPLEGVDGSRVK---AVAVCHTDTSQWNPKHLAFQVLNVQPGTVPVC 367
+YCH +VY L + S+ K VA+CH DT+ W+P H AF L PG + VC
Sbjct: 395 YYCHSVPKVRVYQADLLDPE-SKAKINHGVAICHLDTTAWSPTHGAFMALGSGPGRIEVC 571
Query: 368 HFLPQDHVVW 377
H++ ++ + W
Sbjct: 572 HWIFENDLTW 601
>TC207132 similar to UP|EA92_VICFA (P21747) Embryonic abundant protein USP92
precursor, partial (51%)
Length = 847
Score = 135 bits (340), Expect = 3e-32
Identities = 80/215 (37%), Positives = 117/215 (54%), Gaps = 3/215 (1%)
Frame = +3
Query: 167 FLEKDLHQGTKLNLQFTKTNSNDDAKFLPKEVANSIPFSS---SKVENILNLFSIKEGTE 223
F E DLH ++L K N ++ S PF + +KVE NL +
Sbjct: 258 FFEHDLHPRKIMHLGLHKHNDTKNSTISSWARTTSQPFGAWLHAKVEERYNLDEV----- 422
Query: 224 ESEIVKNTISECEENGIKGEEKLCVTSLESMVDFTTSKLGNNVEAVSTEVKKESSDLQEY 283
C + KGEEK C TSL+SM+ F SKLG N++A+S+ + D +Y
Sbjct: 423 -----------CGKAAAKGEEKFCATSLQSMMGFAISKLGKNIKAISSSF---AQDHDQY 560
Query: 284 VMAKGVKKLGEKNKAVVCHKESYPYAVFYCHKTDSTKVYSVPLEGVDGSRVKAVAVCHTD 343
V+ + V K+GEK AV+CH+ ++ VFYCH+ ++T Y VPL DG++ KA+ +CH D
Sbjct: 561 VVEE-VNKIGEK--AVMCHRLNFENVVFYCHQINATTTYMVPLVASDGTKAKALTICHHD 731
Query: 344 TSQWNPKHLAFQVLNVQPGTVPVCHFLPQDHVVWV 378
T +P + ++VL V+ GTVPVCHF+ + WV
Sbjct: 732 TRGMDP-IVVYEVLKVKTGTVPVCHFVGNKAIAWV 833
Score = 28.9 bits (63), Expect = 4.0
Identities = 13/41 (31%), Positives = 22/41 (52%)
Frame = +3
Query: 1 MCVYVVLQLALVATHAALPPEVYWKSKLPTTQMPKAITDLL 41
+ V +L + L A+ + YW S P T++PK + DL+
Sbjct: 72 LIVLALLCVTLKGADASQSAKDYWHSVFPNTRLPKPLWDLV 194
>TC225145 homologue to UP|O24482 (O24482) Sali3-2, complete
Length = 1382
Score = 129 bits (325), Expect = 2e-30
Identities = 78/223 (34%), Positives = 118/223 (51%)
Frame = +3
Query: 156 QLHDKPNVALFFLEKDLHQGTKLNLQFTKTNSNDDAKFLPKEVANSIPFSSSKVENILNL 215
Q+ D FF ++DLH G + +QFTK P + + +++
Sbjct: 369 QIDDTQYPKTFFYKEDLHPGKTMKVQFTKR---------PYAQPYGVYTWLTDIKD---- 509
Query: 216 FSIKEGTEESEIVKNTISECEENGIKGEEKLCVTSLESMVDFTTSKLGNNVEAVSTEVKK 275
+ KEG EI ++ +GEEK C SL +++ F SKLG N++ +S+
Sbjct: 510 -TSKEGYSFEEIC------IKKEAFEGEEKFCAKSLGTVIGFAISKLGKNIQVLSSSFVN 668
Query: 276 ESSDLQEYVMAKGVKKLGEKNKAVVCHKESYPYAVFYCHKTDSTKVYSVPLEGVDGSRVK 335
+ QE +GV+ LG+K AV+CH ++ AVFYCHK T + VPL DG++ +
Sbjct: 669 K----QEQYTVEGVQNLGDK--AVMCHGLNFRTAVFYCHKVRETTAFMVPLVAGDGTKTQ 830
Query: 336 AVAVCHTDTSQWNPKHLAFQVLNVQPGTVPVCHFLPQDHVVWV 378
A+AVCH+DTS N H+ +++ V PGT PVCHFL ++WV
Sbjct: 831 ALAVCHSDTSGMN-HHMLHELMGVDPGTNPVCHFLGSKAILWV 956
Score = 33.9 bits (76), Expect = 0.12
Identities = 16/40 (40%), Positives = 24/40 (60%), Gaps = 3/40 (7%)
Frame = +3
Query: 5 VVLQLALVA---THAALPPEVYWKSKLPTTQMPKAITDLL 41
++ LAL HA+LP E YW++ P T +P A+ +LL
Sbjct: 207 ILFSLALAGESHVHASLPEEDYWEAVWPNTPIPTALRELL 326
>TC214787 UP|Q06765 (Q06765) ADR6 protein (Sali5-4a protein), complete
Length = 1150
Score = 124 bits (310), Expect = 9e-29
Identities = 76/223 (34%), Positives = 118/223 (52%)
Frame = +3
Query: 156 QLHDKPNVALFFLEKDLHQGTKLNLQFTKTNSNDDAKFLPKEVANSIPFSSSKVENILNL 215
Q+ + FF ++DLH G + +QF+K PF L
Sbjct: 240 QIEETQYPKTFFYKEDLHPGKTMKVQFSKP-----------------PFQQPWGVGTW-L 365
Query: 216 FSIKEGTEESEIVKNTISECEENGIKGEEKLCVTSLESMVDFTTSKLGNNVEAVSTEVKK 275
IK+ T+E + + E I+GEEK C SL +++ F SKLG N++ +S+
Sbjct: 366 KEIKDTTKEGYSFEELCIKKE--AIEGEEKFCAKSLGTVIGFAISKLGKNIQVLSSSFVN 539
Query: 276 ESSDLQEYVMAKGVKKLGEKNKAVVCHKESYPYAVFYCHKTDSTKVYSVPLEGVDGSRVK 335
+ Q+ +GV+ LG+K AV+CH+ ++ AVFYCH+ T + VPL DG++ +
Sbjct: 540 K----QDQYTVEGVQNLGDK--AVMCHRLNFRTAVFYCHEVRETTAFMVPLVAGDGTKTQ 701
Query: 336 AVAVCHTDTSQWNPKHLAFQVLNVQPGTVPVCHFLPQDHVVWV 378
A+A+CH++TS N + L Q++ V PGT PVCHFL ++WV
Sbjct: 702 ALAICHSNTSGMNHQML-HQLMGVDPGTNPVCHFLGSKAILWV 827
>TC234549 weakly similar to UP|O65009 (O65009) BURP domain containing
protein, partial (14%)
Length = 791
Score = 119 bits (297), Expect = 3e-27
Identities = 61/173 (35%), Positives = 99/173 (56%), Gaps = 7/173 (4%)
Frame = +1
Query: 213 LNLFSIKEGTEESEIVKNTISECEENGIKGEEKLCVTSLESMVDFTTSKLGNNVE--AVS 270
L LFSI E + + +++T+ +C I GE K+C TSLESM++F +G + ++
Sbjct: 25 LQLFSISEDSPXANAMRDTLDQCXXEPITGETKICATSLESMLEFVGKIIGLETKHNIIT 204
Query: 271 TEVKKESS-DLQEYVMAKGVKKLGEKNKAVVCHKESYPYAVFYCHK-TDSTKVYSVPL-- 326
T + S LQ++ + + + + +K V CH YPYA++YCH +KV+ V L
Sbjct: 205 TTLPTASGVPLQKFTILEVSEDINA-SKWVACHPLPYPYAIYYCHFIATGSKVFKVSLGS 381
Query: 327 -EGVDGSRVKAVAVCHTDTSQWNPKHLAFQVLNVQPGTVPVCHFLPQDHVVWV 378
D +++A+ +CH DTS W+P H+ F+ L ++PG VCHF H++WV
Sbjct: 382 ENNGDDDKIEALGICHLDTSDWSPNHIIFRQLGIKPGKDSVCHFFTIKHLMWV 540
>TC206937 similar to UP|Q40161 (Q40161) Polygalacturonase isoenzyme 1 beta
subunit precursor , partial (23%)
Length = 537
Score = 117 bits (292), Expect = 1e-26
Identities = 58/173 (33%), Positives = 91/173 (52%), Gaps = 2/173 (1%)
Frame = +2
Query: 207 SKVENILNLFSIKEGTEESEIVKNTISECEENGIKGEEKLCVTSLESMVDFTTSKLGNNV 266
SK++ + +F + +++K+++ ECE GE K CV SLE M+DF TS LG NV
Sbjct: 5 SKIDELKQVFKASDNGSMEKMMKDSLEECERAPSSGETKRCVGSLEDMIDFATSVLGRNV 184
Query: 267 EAVSTEVKKESSDLQEYVMAKGVKKLGEKNKAVVCHKESYPYAVFYCHKTDSTKVYSVPL 326
+T+ S +G+ G+ ++V CH+ +PY ++YCH +VY L
Sbjct: 185 AVRTTQNVNGSKKSVVVGPVRGING-GKVTQSVSCHQSLFPYLLYYCHAVPKVRVYEADL 361
Query: 327 --EGVDGSRVKAVAVCHTDTSQWNPKHLAFQVLNVQPGTVPVCHFLPQDHVVW 377
+ VA+CH DTS W+P H AF L PG + VCH++ ++ + W
Sbjct: 362 LDPKTKAKINRGVAICHLDTSDWSPTHGAFLSLGSVPGRIEVCHWIFENDMAW 520
>BQ273716 weakly similar to SP|Q08298|RD22_ Dehydration-responsive protein
RD22 precursor. [Mouse-ear cress] {Arabidopsis
thaliana}, partial (13%)
Length = 419
Score = 113 bits (283), Expect = 1e-25
Identities = 44/84 (52%), Positives = 59/84 (69%)
Frame = +1
Query: 297 KAVVCHKESYPYAVFYCHKTDSTKVYSVPLEGVDGSRVKAVAVCHTDTSQWNPKHLAFQV 356
K +VCH YPY VF CH+ +T + +PLEG DG+RVKA AVCH DTS+W+P H+ Q+
Sbjct: 67 KVIVCHPMDYPYVVFMCHEISNTTAHFMPLEGEDGTRVKAAAVCHKDTSEWDPNHVFLQM 246
Query: 357 LNVQPGTVPVCHFLPQDHVVWVSK 380
L +PG PVCH P+ H++W +K
Sbjct: 247 LKTKPGAAPVCHIFPEGHLLWFAK 318
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.312 0.131 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,436,661
Number of Sequences: 63676
Number of extensions: 222991
Number of successful extensions: 1426
Number of sequences better than 10.0: 134
Number of HSP's better than 10.0 without gapping: 1166
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1279
length of query: 380
length of database: 12,639,632
effective HSP length: 99
effective length of query: 281
effective length of database: 6,335,708
effective search space: 1780333948
effective search space used: 1780333948
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148532.8


