

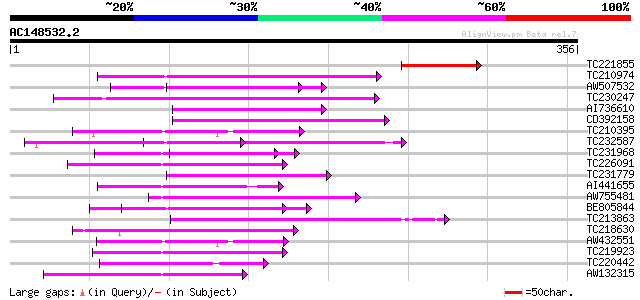
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148532.2 - phase: 0
(356 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC221855 89 3e-18
TC210974 similar to UP|Q6K9W7 (Q6K9W7) Pentatricopeptide (PPR) r... 59 3e-09
AW507532 58 7e-09
TC230247 weakly similar to GB|AAP88326.1|32815835|BT009692 At1g6... 58 7e-09
AI736610 57 2e-08
CD392158 55 5e-08
TC210395 similar to UP|Q9FGR7 (Q9FGR7) Similarity to salt-induci... 55 5e-08
TC232587 similar to UP|Q9LME0 (Q9LME0) F14D16.2, partial (17%) 55 6e-08
TC231968 52 3e-07
TC226091 similar to UP|Q8L844 (Q8L844) Crp1 protein-like, partia... 52 5e-07
TC231779 weakly similar to UP|Q9SGV7 (Q9SGV7) F1N19.15, partial ... 52 5e-07
AI441655 51 7e-07
AW755481 similar to GP|2244996|emb salt-inducible protein homolo... 51 7e-07
BE805844 49 3e-06
TC213863 similar to UP|Q9FKC3 (Q9FKC3) Similarity to salt-induci... 48 6e-06
TC218630 weakly similar to PIR|D86391|D86391 T1K7.17 protein - A... 48 8e-06
AW432551 similar to GP|9759030|dbj contains similarity to salt-i... 48 8e-06
TC219923 similar to UP|Q9FMF6 (Q9FMF6) Gb|AAF15929.1, partial (10%) 47 1e-05
TC220442 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding p... 46 3e-05
AW132315 weakly similar to GP|6630464|gb| F23N19.4 {Arabidopsis ... 45 4e-05
>TC221855
Length = 663
Score = 89.0 bits (219), Expect = 3e-18
Identities = 39/50 (78%), Positives = 48/50 (96%)
Frame = +2
Query: 247 SQIHCGIVTMYAMLDRLDEVELSVGRMLKQGMSFTSSDDVEKVICSYFRK 296
++IHCGIV MYAM+DRLD++E +VGRMLKQG+SFTS+DDVEKVICSYFR+
Sbjct: 2 TRIHCGIVKMYAMVDRLDDIEFAVGRMLKQGLSFTSADDVEKVICSYFRR 151
>TC210974 similar to UP|Q6K9W7 (Q6K9W7) Pentatricopeptide (PPR)
repeat-containing protein-like, partial (3%)
Length = 637
Score = 59.3 bits (142), Expect = 3e-09
Identities = 41/178 (23%), Positives = 82/178 (46%)
Frame = +2
Query: 56 VFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAF 115
VF M G++PT +N L+ A S + A ++F+ M + Y PD +Y +SA+
Sbjct: 2 VFEEMLDAGIRPTRKAYNILLDAFSISGMVEQAQTVFKSMRR-DRYFPDLCSYTTMLSAY 178
Query: 116 SKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPNV 175
+ +++ ++ G P++ T+ ++I G + E+ + +EEM + + N
Sbjct: 179 INADDMEGAEKFFKRLIQDGFEPNVVTYGTLIKGYAKINDLEMVMKKYEEMLMRGIKANQ 358
Query: 176 TILESMLKGFCSQKSLCRANEFFKFVLDNRWQISETMAAMLVVLYHEQGQVEKMEELL 233
TIL +++ + A +FK + N + +L+ L + E+ EL+
Sbjct: 359 TILTTIMDAYGKSGDFDSAVHWFKEMESNGIPPDQKAKNVLLSLAKTDEEREEANELV 532
Score = 37.0 bits (84), Expect = 0.014
Identities = 22/112 (19%), Positives = 47/112 (41%)
Frame = +2
Query: 64 GVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDA 123
G +P + + +LI +D+ +E M K + + A+ KSG+ D+
Sbjct: 236 GFEPNVVTYGTLIKGYAKINDLEMVMKKYEEM-LMRGIKANQTILTTIMDAYGKSGDFDS 412
Query: 124 MLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPNV 175
+ W+ ++ G+ PD + ++S + E A+ + + +P V
Sbjct: 413 AVHWFKEMESNGIPPDQKAKNVLLSLAKTDEEREEANELVVHFSENSSLPKV 568
>AW507532
Length = 396
Score = 57.8 bits (138), Expect = 7e-09
Identities = 30/101 (29%), Positives = 55/101 (53%)
Frame = +3
Query: 99 ENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEI 158
E P HT+N + S +D + +Y K+ G+ PD+ T+ ++I+G K +
Sbjct: 30 EGVDPTRHTFNILLWGMFLSLRLDTAVRFYEDMKSRGILPDVVTYNTLINGYFRFKKVDE 209
Query: 159 ADRVFEEMKISEMIPNVTILESMLKGFCSQKSLCRANEFFK 199
A+++F EMK +++PNV +MLKG+ + + A + F+
Sbjct: 210 AEKLFVEMKGRDIVPNVISFTTMLKGYVAAGRIDDALKVFE 332
Score = 57.0 bits (136), Expect = 1e-08
Identities = 38/121 (31%), Positives = 59/121 (48%)
Frame = +3
Query: 64 GVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDA 123
GV PT FN L+ S + TA +E M+S PD TYN I+ + + VD
Sbjct: 33 GVDPTRHTFNILLWGMFLSLRLDTAVRFYEDMKS-RGILPDVVTYNTLINGYFRFKKVDE 209
Query: 124 MLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLK 183
+ K + P++ +F +++ G V + + A +VFEEMK + PNV ++L
Sbjct: 210 AEKLFVEMKGRDIVPNVISFTTMLKGYVAAGRIDDALKVFEEMKGCGVKPNVVTFSTLLP 389
Query: 184 G 184
G
Sbjct: 390 G 392
>TC230247 weakly similar to GB|AAP88326.1|32815835|BT009692 At1g61870
{Arabidopsis thaliana;} , partial (56%)
Length = 1291
Score = 57.8 bits (138), Expect = 7e-09
Identities = 39/205 (19%), Positives = 92/205 (44%)
Frame = +3
Query: 28 KENVKDHGRFSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVT 87
+ ++++ S I L G+ + I FT E + + NSL+ A L + +
Sbjct: 300 RPDLRNEKFLSHAIVLYGQANMLDHAIRTFT--EDLPSPRSVKTLNSLLFAALLAKNYKE 473
Query: 88 AYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVI 147
++ + + +P+ TYN I AF++SG+ ++ + + + P++ T + +
Sbjct: 474 LTRIYLEFPKTYSIQPNLDTYNTVIKAFAESGSTSSVYSVLAEMDKNNIAPNVTTLNNSL 653
Query: 148 SGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLKGFCSQKSLCRANEFFKFVLDNRWQ 207
SG K ++ +V + M+ + P+++ ++ C K A + ++ N +
Sbjct: 654 SGFYREKKFDDVGKVLKLMEKYSVFPSISTYNVRIQSLCKLKRSSEAKALLEGMVCNGRK 833
Query: 208 ISETMAAMLVVLYHEQGQVEKMEEL 232
+ A L+ + ++G +E+ + L
Sbjct: 834 PNSVSYACLIHGFCKEGDLEEAKRL 908
>AI736610
Length = 411
Score = 56.6 bits (135), Expect = 2e-08
Identities = 29/97 (29%), Positives = 54/97 (54%)
Frame = +2
Query: 103 PDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRV 162
P HT+N + S +D + +Y K+ G+ PD+ T+ ++I+G K + A+++
Sbjct: 119 PTRHTFNILLWGMFLSLRLDTAVRFYEDMKSRGILPDVVTYNTLINGYFRFKKVDEAEKL 298
Query: 163 FEEMKISEMIPNVTILESMLKGFCSQKSLCRANEFFK 199
F EMK +++PNV +MLKG+ + + A + F+
Sbjct: 299 FVEMKGRDIVPNVISFTTMLKGYVAAGRIDDALKVFE 409
>CD392158
Length = 704
Score = 55.1 bits (131), Expect = 5e-08
Identities = 30/136 (22%), Positives = 63/136 (46%)
Frame = -2
Query: 103 PDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRV 162
P TYN I +K G + + + GL PD+ T+ +++ G + A ++
Sbjct: 676 PVLITYNTVIDGLTKVGKTEYAVELLEEMRRKGLKPDIITYSTLLRGLGREGKVDEAIKI 497
Query: 163 FEEMKISEMIPNVTILESMLKGFCSQKSLCRANEFFKFVLDNRWQISETMAAMLVVLYHE 222
F +M+ + P+ +++ G C + RA +F ++++ + +E +L+ +
Sbjct: 496 FHDMEGLSIKPSAVTYNAIMLGLCKAQQTSRAIDFLAYMVEKGCKPTEATYTILIEGIAD 317
Query: 223 QGQVEKMEELLETITS 238
+G E+ ELL + S
Sbjct: 316 EGLAEEALELLNELCS 269
Score = 37.0 bits (84), Expect = 0.014
Identities = 25/105 (23%), Positives = 48/105 (44%), Gaps = 1/105 (0%)
Frame = -2
Query: 22 ILEKLLKENVK-DHGRFSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACL 80
+LE++ ++ +K D +S L+ G+ V I +F MEG+ +KP+++ +N+++
Sbjct: 604 LLEEMRRKGLKPDIITYSTLLRGLGREGKVDEAIKIFHDMEGLSIKPSAVTYNAIMLGLC 425
Query: 81 SSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDAML 125
+ A M + KP TY I + G + L
Sbjct: 424 KAQQTSRAIDFLAYM-VEKGCKPTEATYTILIEGIADEGLAEEAL 293
>TC210395 similar to UP|Q9FGR7 (Q9FGR7) Similarity to salt-inducible protein,
partial (33%)
Length = 954
Score = 55.1 bits (131), Expect = 5e-08
Identities = 43/151 (28%), Positives = 67/151 (43%), Gaps = 5/151 (3%)
Frame = +1
Query: 40 LIFLCGKLKNVQ--LGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMES 97
LI GK KN+ + F M+ VGVKPTS + +LI A S AY+ FE M+
Sbjct: 43 LIIAYGKQKNMSDMAAADAFLKMKKVGVKPTSQSYTALIHAYSVSGLHEKAYAAFENMQ- 219
Query: 98 SENYKPDFHTYNNFISAFSKSGNVDAMLAWYS---AKKATGLGPDLQTFESVISGCVNSK 154
+E KP TY ++AF +G+ ++ + ++K G G TF ++ G
Sbjct: 220 NEGIKPSIETYTTLLNAFRHAGDAQTLMEIWKLMISEKVEGTG---ATFNILVDGFAKQG 390
Query: 155 NYEIADRVFEEMKISEMIPNVTILESMLKGF 185
+ A V E + P V ++ +
Sbjct: 391 LFMEAREVISEFGKVGLKPTVVTYNMLINAY 483
>TC232587 similar to UP|Q9LME0 (Q9LME0) F14D16.2, partial (17%)
Length = 516
Score = 54.7 bits (130), Expect = 6e-08
Identities = 35/145 (24%), Positives = 72/145 (49%), Gaps = 6/145 (4%)
Frame = +3
Query: 10 LIINIL-----IQVLEWILEKLLKEN-VKDHGRFSELIFLCGKLKNVQLGINVFTSMEGV 63
+IIN L + W+ ++++ V + ++ +I L K +N ++ + ++ M+
Sbjct: 81 VIINCLGKAGNLAAAHWLFCEMVEHGCVPNLVTYNIMIALQAKARNYEMALKLYHDMQNA 260
Query: 64 GVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDA 123
G +P + ++ ++ A + A S+F M+ +N+ PD Y + + K+GNV+
Sbjct: 261 GFQPDKVTYSIVMEALGHCGYLEEAESVFVEMQQ-KNWVPDEPVYGLLVDLWGKAGNVEK 437
Query: 124 MLAWYSAKKATGLGPDLQTFESVIS 148
WY A GL P++ T S++S
Sbjct: 438 ASEWYQAMLNAGLLPNVPTCNSLLS 512
Score = 53.9 bits (128), Expect = 1e-07
Identities = 39/165 (23%), Positives = 78/165 (46%)
Frame = +3
Query: 85 IVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFE 144
I A S+++ M+ + PD TY+ I+ K+GN+ A + G P+L T+
Sbjct: 9 IDVAMSMYKRMQEA-GLSPDTFTYSVIINCLGKAGNLAAAHWLFCEMVEHGCVPNLVTYN 185
Query: 145 SVISGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLKGFCSQKSLCRANEFFKFVLDN 204
+I+ ++NYE+A +++ +M+ + P+ +++ L A F +
Sbjct: 186 IMIALQAKARNYEMALKLYHDMQNAGFQPDKVTYSIVMEALGHCGYLEEAESVFVEMQQK 365
Query: 205 RWQISETMAAMLVVLYHEQGQVEKMEELLETITSYPIDSGVLSQI 249
W E + +LV L+ + G VEK E + + +++G+L +
Sbjct: 366 NWVPDEPVYGLLVDLWGKAGNVEKASEWYQAM----LNAGLLPNV 488
Score = 40.4 bits (93), Expect = 0.001
Identities = 31/146 (21%), Positives = 64/146 (43%)
Frame = +3
Query: 37 FSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIME 96
+S +I GK N+ +F M G P + +N +I+ + + A L+ M+
Sbjct: 75 YSVIINCLGKAGNLAAAHWLFCEMVEHGCVPNLVTYNIMIALQAKARNYEMALKLYHDMQ 254
Query: 97 SSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNY 156
++ ++PD TY+ + A G ++ + + + PD + ++ + N
Sbjct: 255 NA-GFQPDKVTYSIVMEALGHCGYLEEAESVFVEMQQKNWVPDEPVYGLLVDLWGKAGNV 431
Query: 157 EIADRVFEEMKISEMIPNVTILESML 182
E A ++ M + ++PNV S+L
Sbjct: 432 EKASEWYQAMLNAGLLPNVPTCNSLL 509
>TC231968
Length = 610
Score = 52.4 bits (124), Expect = 3e-07
Identities = 30/116 (25%), Positives = 56/116 (47%)
Frame = +1
Query: 54 INVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFIS 113
++VF S++ G P ++ +N+ I+ + + F+ M + + +PD TY I
Sbjct: 25 LDVFASIKDTGFVPDTVSYNTXINGLRKAGRXDMCFVYFKEM-TEKGVEPDLLTYTAIIE 201
Query: 114 AFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKIS 169
F +SGNV+ L + K G+ P + + S+I + E+A + EE+ S
Sbjct: 202 IFGRSGNVEESLKCFREMKLKGVLPSIYIYRSLIHNLNKTGKVELATELLEELNSS 369
Score = 50.1 bits (118), Expect = 2e-06
Identities = 24/82 (29%), Positives = 43/82 (52%)
Frame = +1
Query: 101 YKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIAD 160
+ PD +YN I+ K+G D ++ G+ PDL T+ ++I S N E +
Sbjct: 58 FVPDTVSYNTXINGLRKAGRXDMCFVYFKEMTEKGVEPDLLTYTAIIEIFGRSGNVEESL 237
Query: 161 RVFEEMKISEMIPNVTILESML 182
+ F EMK+ ++P++ I S++
Sbjct: 238 KCFREMKLKGVLPSIYIYRSLI 303
>TC226091 similar to UP|Q8L844 (Q8L844) Crp1 protein-like, partial (23%)
Length = 849
Score = 51.6 bits (122), Expect = 5e-07
Identities = 33/138 (23%), Positives = 62/138 (44%)
Frame = +2
Query: 37 FSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIME 96
++ L+ + GK I ++ G KPTS ++N+LI+A A + F +M
Sbjct: 29 YTTLVDVYGKSGRFSDAIECLEVLKSTGFKPTSTMYNALINAYAQRGLSELAVNAFRLM- 205
Query: 97 SSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNY 156
++E P N+ I+AF + A K + PD+ T+ +++ + + +
Sbjct: 206 TTEGLTPSLLALNSLINAFGEDRRDAEAFAVLQYMKENNIEPDVVTYTTLMKALIRVEKF 385
Query: 157 EIADRVFEEMKISEMIPN 174
+ V+EEM S P+
Sbjct: 386 QKVPAVYEEMVTSGCTPD 439
>TC231779 weakly similar to UP|Q9SGV7 (Q9SGV7) F1N19.15, partial (7%)
Length = 620
Score = 51.6 bits (122), Expect = 5e-07
Identities = 28/104 (26%), Positives = 50/104 (47%)
Frame = +1
Query: 99 ENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEI 158
+N PD TYN+ I KSG + + L + G D+ T+ S++ ++N++
Sbjct: 16 KNMVPDTVTYNSLIDGLCKSGRITSALNLMNEMHHRGQPADVVTYTSLLDALCKNQNHDK 195
Query: 159 ADRVFEEMKISEMIPNVTILESMLKGFCSQKSLCRANEFFKFVL 202
A +F +MK + P + +++ G C L A E F+ +L
Sbjct: 196 ATALFMKMKKRRIQPTMYTYTALIDGLCKGGRLKNAQELFQHLL 327
Score = 40.4 bits (93), Expect = 0.001
Identities = 36/160 (22%), Positives = 63/160 (38%)
Frame = +1
Query: 24 EKLLKENVKDHGRFSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSH 83
E L K V D ++ LI K + +N+ M G + + SL+ A +
Sbjct: 4 EMLHKNMVPDTVTYNSLIDGLCKSGRITSALNLMNEMHHRGQPADVVTYTSLLDALCKNQ 183
Query: 84 DIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTF 143
+ A +LF M+ +P +TY I K G + + G D+ T+
Sbjct: 184 NHDKATALFMKMKK-RRIQPTMYTYTALIDGLCKGGRLKNAQELFQHLLVKGCCIDVWTY 360
Query: 144 ESVISGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLK 183
+ISG + A + +M+ + IPN E +++
Sbjct: 361 TVMISGLCKEGMLDEALAMKSKMEDNGCIPNAVTFEIIIR 480
>AI441655
Length = 405
Score = 51.2 bits (121), Expect = 7e-07
Identities = 34/117 (29%), Positives = 56/117 (47%)
Frame = +2
Query: 56 VFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAF 115
VF ++ G KPT + + +L++A + +L + + KPD N I+AF
Sbjct: 38 VFNNLTEEGHKPTLITYTTLVAALTRQKRFKSIPALLSKV-ADNGMKPDSILLNAMINAF 214
Query: 116 SKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMI 172
S+SG VD + + K P T+ ++I G + IA R +E MK+ EM+
Sbjct: 215 SESGKVDEAMKIFQKMKEX*CKPTTSTYNTLIKG------FGIAGRPYESMKLLEMM 367
>AW755481 similar to GP|2244996|emb salt-inducible protein homolog
{Arabidopsis thaliana}, partial (17%)
Length = 435
Score = 51.2 bits (121), Expect = 7e-07
Identities = 37/133 (27%), Positives = 62/133 (45%)
Frame = +2
Query: 88 AYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVI 147
A FE M SS +PD TY+ I A+ ++GN+D L Y + D TF ++I
Sbjct: 17 AVEWFEKM-SSFGCEPDDVTYSAMIDAYGRAGNIDMALRLYDRARTEKWRLDTVTFSTLI 193
Query: 148 SGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLKGFCSQKSLCRANEFFKFVLDNRWQ 207
+ NY+ V++EMK+ + PN+ I ++L K +A + + +N +
Sbjct: 194 KMYGLAGNYDGCLNVYQEMKVLGVKPNMVIYNTLLDAMGRAKRPWQAKSIYTEMTNNGFS 373
Query: 208 ISETMAAMLVVLY 220
+ A L+ Y
Sbjct: 374 PNWVTYASLLRAY 412
Score = 40.4 bits (93), Expect = 0.001
Identities = 23/81 (28%), Positives = 45/81 (55%)
Frame = +2
Query: 37 FSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIME 96
FS LI + G N +NV+ M+ +GVKP +++N+L+ A + A S++ M
Sbjct: 179 FSTLIKMYGLAGNYDGCLNVYQEMKVLGVKPNMVIYNTLLDAMGRAKRPWQAKSIYTEM- 355
Query: 97 SSENYKPDFHTYNNFISAFSK 117
++ + P++ TY + + A+ +
Sbjct: 356 TNNGFSPNWVTYASLLRAYGR 418
>BE805844
Length = 429
Score = 48.9 bits (115), Expect = 3e-06
Identities = 29/119 (24%), Positives = 58/119 (48%)
Frame = +3
Query: 71 VFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSA 130
VF+ +++ + ++ + + M+SS +PD H YN I F K +D +A +
Sbjct: 3 VFSRILANYRDKGEWQKSFQVLKDMKSS-GVQPDRHFYNVMIDTFGKYNCLDHAMATFER 179
Query: 131 KKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIPNVTILESMLKGFCSQK 189
+ G+ PD+ T+ ++I S +++A+ +F EM+ P +T M+ Q+
Sbjct: 180 MLSEGIPPDIVTWNTLIDCHCKSGRHDMAEELFSEMQQRGYSPCITTYNIMINSMGEQQ 356
Score = 45.1 bits (105), Expect = 5e-05
Identities = 32/124 (25%), Positives = 51/124 (40%)
Frame = +3
Query: 51 QLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNN 110
Q V M+ GV+P +N +I + + A + FE M SE PD T+N
Sbjct: 48 QKSFQVLKDMKSSGVQPDRHFYNVMIDTFGKYNCLDHAMATFERM-LSEGIPPDIVTWNT 224
Query: 111 FISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISE 170
I KSG D +S + G P + T+ +I+ + +E +M+
Sbjct: 225 LIDCHCKSGRHDMAEELFSEMQQRGYSPCITTYNIMINSMGEQQRWEQVTAFLSKMQSQG 404
Query: 171 MIPN 174
+ PN
Sbjct: 405 LQPN 416
Score = 37.7 bits (86), Expect = 0.008
Identities = 28/111 (25%), Positives = 48/111 (43%)
Frame = +3
Query: 33 DHGRFSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLF 92
D ++ +I GK + + F M G+ P + +N+LI S A LF
Sbjct: 99 DRHFYNVMIDTFGKYNCLDHAMATFERMLSEGIPPDIVTWNTLIDCHCKSGRHDMAEELF 278
Query: 93 EIMESSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTF 143
M+ Y P TYN I++ + + + A+ S ++ GL P+ T+
Sbjct: 279 SEMQQ-RGYSPCITTYNIMINSMGEQQRWEQVTAFLSKMQSQGLQPNSITY 428
>TC213863 similar to UP|Q9FKC3 (Q9FKC3) Similarity to salt-inducible protein,
partial (41%)
Length = 821
Score = 48.1 bits (113), Expect = 6e-06
Identities = 43/176 (24%), Positives = 75/176 (42%), Gaps = 1/176 (0%)
Frame = +2
Query: 102 KPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADR 161
+PD T N+ + AF G ++ M Y + G+ P++QTF ++ +++Y+
Sbjct: 185 QPDVWTMNSTLRAFGNIGQIETMEKCYEKFQNAGIQPNVQTFNILLDSYGKAQDYKKMSA 364
Query: 162 VFEEMKISEMIPNVTILESMLKGFCSQKSLCRANEFFKFVLDNRWQISETMAAMLVVLYH 221
V E M+ + ++ F L + F+ + R + S LV Y
Sbjct: 365 VMEYMQKYHYSWTIVTFNIVIDAFGKAGDLKQMEYLFRLMRSERIKPSCVTLCSLVRAYA 544
Query: 222 EQGQVEKMEELLETI-TSYPIDSGVLSQIHCGIVTMYAMLDRLDEVELSVGRMLKQ 276
G +K +L + SY + V +C +V YA L L E++ V M+KQ
Sbjct: 545 RAGNSDKFSGVLRFVENSYVLLDTVF--FNC-LVDAYARLGCLAEMK-GVLEMMKQ 700
>TC218630 weakly similar to PIR|D86391|D86391 T1K7.17 protein - Arabidopsis
thaliana {Arabidopsis thaliana;} , partial (52%)
Length = 1513
Score = 47.8 bits (112), Expect = 8e-06
Identities = 33/146 (22%), Positives = 68/146 (45%), Gaps = 4/146 (2%)
Frame = +1
Query: 40 LIFLCGKLKNVQLGINVFTSMEGVG-VKP---TSLVFNSLISACLSSHDIVTAYSLFEIM 95
L+ C K K + NV+ +E + +P + N +I C + D+ AY FE +
Sbjct: 487 LVVACSK-KGFETLDNVYFQLENLNRAEPPYKSVAALNCVILGCANIWDLDRAYQTFESI 663
Query: 96 ESSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKN 155
S+ PD H+YN + AF K + + GL P+ +++ ++ + +++
Sbjct: 664 GSTFGLIPDIHSYNGLMYAFGKLKKTHEATRVFEHLVSLGLKPNAKSYSLLVDAHLINRD 843
Query: 156 YEIADRVFEEMKISEMIPNVTILESM 181
+ A V ++M+ + P+ +L+ +
Sbjct: 844 VKSALAVIDDMRAAGYEPSKEVLKKV 921
Score = 37.4 bits (85), Expect = 0.010
Identities = 19/73 (26%), Positives = 40/73 (54%)
Frame = +1
Query: 31 VKDHGRFSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYS 90
+ D ++ L++ GKLK VF + +G+KP + ++ L+ A L + D+ +A +
Sbjct: 682 IPDIHSYNGLMYAFGKLKKTHEATRVFEHLVSLGLKPNAKSYSLLVDAHLINRDVKSALA 861
Query: 91 LFEIMESSENYKP 103
+ + M ++ Y+P
Sbjct: 862 VIDDMRAA-GYEP 897
>AW432551 similar to GP|9759030|dbj contains similarity to salt-inducible
protein~gene_id:K6A12.14 {Arabidopsis thaliana}, partial
(19%)
Length = 443
Score = 47.8 bits (112), Expect = 8e-06
Identities = 36/124 (29%), Positives = 55/124 (44%), Gaps = 3/124 (2%)
Frame = +3
Query: 55 NVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISA 114
+ F M+ VG+KPT + +LI A S AY+ FE M+ SE KP TY +
Sbjct: 21 DAFLKMKKVGIKPTLHSYTALIHAYSVSGLHEKAYTAFENMQ-SEGIKPSIETYTTLLDV 197
Query: 115 FSKSGNVDAMLAWYS---AKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEM 171
F ++G+ ++ + ++K G G TF ++ G Y A V E +
Sbjct: 198 FRRAGDAQTLMKIWKLMMSEKVEGTG---VTFNILVDGFAKQGLYMEAREVISEFGKVGL 368
Query: 172 IPNV 175
P V
Sbjct: 369 QPTV 380
>TC219923 similar to UP|Q9FMF6 (Q9FMF6) Gb|AAF15929.1, partial (10%)
Length = 1238
Score = 47.0 bits (110), Expect = 1e-05
Identities = 33/122 (27%), Positives = 55/122 (45%)
Frame = +1
Query: 53 GINVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFI 112
G+ +F M G G+ PT + N LIS + + A + M PD TYN+ I
Sbjct: 139 GLGLFEEMLGKGIFPTIISCNILISGLCRTGKVNDALKFLQDM-IHRGLTPDIVTYNSLI 315
Query: 113 SAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMI 172
+ K G+V ++ ++ G+ PD T+ ++IS + + A + + S I
Sbjct: 316 NGLCKMGHVQEASNLFNKLQSEGIRPDAITYNTLISRHCHEGMFNDACLLLYKGVDSGFI 495
Query: 173 PN 174
PN
Sbjct: 496 PN 501
Score = 31.2 bits (69), Expect = 0.75
Identities = 15/59 (25%), Positives = 34/59 (57%)
Frame = +1
Query: 19 LEWILEKLLKENVKDHGRFSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLIS 77
L+++ + + + D ++ LI K+ +VQ N+F ++ G++P ++ +N+LIS
Sbjct: 247 LKFLQDMIHRGLTPDIVTYNSLINGLCKMGHVQEASNLFNKLQSEGIRPDAITYNTLIS 423
>TC220442 weakly similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding protein-like,
partial (25%)
Length = 706
Score = 45.8 bits (107), Expect = 3e-05
Identities = 24/106 (22%), Positives = 50/106 (46%)
Frame = +1
Query: 57 FTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFS 116
FT M+ G+ P S+ F ++++AC + SLFE M S N KP Y +
Sbjct: 79 FTQMQKAGINPNSITFTAILTACSHAGLTEEGKSLFESMSSVYNIKPSMEHYGCMVDLMG 258
Query: 117 KSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRV 162
++G + + ++ + P+ + ++++ C K++E+ +
Sbjct: 259 RAGLLKEAREFI---ESMPVKPNAAIWGALLNACQLHKHFELGKEI 387
>AW132315 weakly similar to GP|6630464|gb| F23N19.4 {Arabidopsis thaliana},
partial (5%)
Length = 411
Score = 45.4 bits (106), Expect = 4e-05
Identities = 31/129 (24%), Positives = 61/129 (47%), Gaps = 1/129 (0%)
Frame = +3
Query: 22 ILEKLLKENVK-DHGRFSELIFLCGKLKNVQLGINVFTSMEGVGVKPTSLVFNSLISACL 80
+L K+ +++ D +S +I K K V + ++ M+ G+ P + + +LI
Sbjct: 18 LLRKIEDRSIRPDVVMYSTIIDSLCKDKLVNEAYDFYSEMDARGIFPDVITYTTLICGXC 197
Query: 81 SSHDIVTAYSLFEIMESSENYKPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDL 140
+ ++ A+SL M +N PD HT++ I A K G V + G+ P++
Sbjct: 198 LASQLMGAFSLLNEM-ILKNINPDVHTFSILIDALCKEGKVKEAKNLLAVMMKEGVKPNV 374
Query: 141 QTFESVISG 149
T+ +++ G
Sbjct: 375 VTYNTLMDG 401
Score = 40.4 bits (93), Expect = 0.001
Identities = 24/133 (18%), Positives = 54/133 (40%)
Frame = +3
Query: 54 INVFTSMEGVGVKPTSLVFNSLISACLSSHDIVTAYSLFEIMESSENYKPDFHTYNNFIS 113
I + +E ++P ++++++I + + AY + M++ + PD TY I
Sbjct: 12 IKLLRKIEDRSIRPDVVMYSTIIDSLCKDKLVNEAYDFYSEMDARGIF-PDVITYTTLIC 188
Query: 114 AFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADRVFEEMKISEMIP 173
+ + + + + PD+ TF +I + A + M + P
Sbjct: 189 GXCLASQLMGAFSLLNEMILKNINPDVHTFSILIDALCKEGKVKEAKNLLAVMMKEGVKP 368
Query: 174 NVTILESMLKGFC 186
NV +++ G+C
Sbjct: 369 NVVTYNTLMDGYC 407
Score = 37.7 bits (86), Expect = 0.008
Identities = 22/101 (21%), Positives = 43/101 (41%)
Frame = +3
Query: 102 KPDFHTYNNFISAFSKSGNVDAMLAWYSAKKATGLGPDLQTFESVISGCVNSKNYEIADR 161
+PD Y+ I + K V+ +YS A G+ PD+ T+ ++I G + A
Sbjct: 48 RPDVVMYSTIIDSLCKDKLVNEAYDFYSEMDARGIFPDVITYTTLICGXCLASQLMGAFS 227
Query: 162 VFEEMKISEMIPNVTILESMLKGFCSQKSLCRANEFFKFVL 202
+ EM + + P+V ++ C + + A ++
Sbjct: 228 LLNEMILKNINPDVHTFSILIDALCKEGKVKEAKNLLAVMM 350
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.137 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,252,292
Number of Sequences: 63676
Number of extensions: 141069
Number of successful extensions: 818
Number of sequences better than 10.0: 89
Number of HSP's better than 10.0 without gapping: 762
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 807
length of query: 356
length of database: 12,639,632
effective HSP length: 98
effective length of query: 258
effective length of database: 6,399,384
effective search space: 1651041072
effective search space used: 1651041072
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148532.2


