

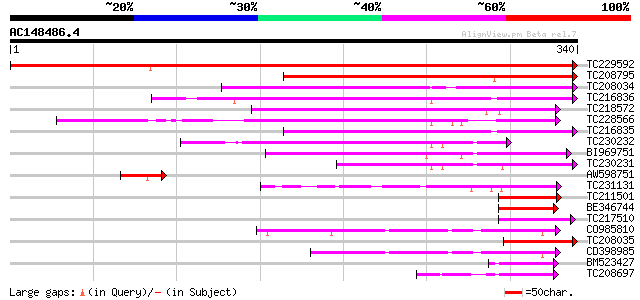
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148486.4 - phase: 0
(340 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC229592 similar to UP|Q9M5Q3 (Q9M5Q3) S-ribonuclease binding pr... 614 e-176
TC208795 similar to UP|Q9SGY4 (Q9SGY4) F20B24.9, partial (28%) 150 1e-36
TC208034 weakly similar to UP|Q9LPJ0 (Q9LPJ0) F6N18.12, partial ... 139 2e-33
TC216836 weakly similar to UP|Q9LPJ0 (Q9LPJ0) F6N18.12, partial ... 129 2e-30
TC218572 similar to UP|Q9FHE4 (Q9FHE4) Arabidopsis thaliana geno... 102 2e-22
TC228566 weakly similar to UP|Q9FHE4 (Q9FHE4) Arabidopsis thalia... 102 2e-22
TC216835 weakly similar to UP|Q9LPJ0 (Q9LPJ0) F6N18.12, partial ... 101 4e-22
TC230232 similar to UP|Q8LAY3 (Q8LAY3) Inhibitor of apoptosis-li... 89 3e-18
BI969751 89 4e-18
TC230231 similar to UP|Q8LAY3 (Q8LAY3) Inhibitor of apoptosis-li... 81 6e-16
AW598751 54 8e-08
TC231131 similar to GB|BAB71852.1|16902294|AB062739 kinesin-rela... 53 2e-07
TC211501 similar to GB|BAB71852.1|16902294|AB062739 kinesin-rela... 51 9e-07
BE346744 weakly similar to GP|21593126|gb| inhibitor of apoptosi... 50 1e-06
TC217510 49 3e-06
CO985810 48 6e-06
TC208035 similar to UP|Q9LPJ0 (Q9LPJ0) F6N18.12, partial (15%) 47 1e-05
CD398985 similar to PIR|T01393|T013 apoptosis inhibitor homolog ... 45 4e-05
BM523427 44 8e-05
TC208697 44 8e-05
>TC229592 similar to UP|Q9M5Q3 (Q9M5Q3) S-ribonuclease binding protein SBP1
(Fragment), partial (91%)
Length = 1267
Score = 614 bits (1584), Expect = e-176
Identities = 302/342 (88%), Positives = 322/342 (93%), Gaps = 2/342 (0%)
Frame = +1
Query: 1 MAFLQDQFQRHYQQQQQPQPQTKSFRNLQTIEGQMSQQMAFYNPTDLQDQSQHPPYIPPF 60
MAF Q FQ+HYQ QQQ QPQTK+FRNLQTI+GQMSQQ+AFYNPTDLQ+QSQHPPYIPPF
Sbjct: 28 MAFPQHHFQQHYQTQQQQQPQTKNFRNLQTIDGQMSQQVAFYNPTDLQEQSQHPPYIPPF 207
Query: 61 HVVGFAPGPVIPADGSDGGVDLH--WNFGLEPERKRLKEQDFLENNSQISSVDFLQPRSV 118
HVVGFAPGPV PADGSDGGVDL WN+GLEPERKRLKEQDFLENNSQISSVDFLQPRSV
Sbjct: 208 HVVGFAPGPVPPADGSDGGVDLQLQWNYGLEPERKRLKEQDFLENNSQISSVDFLQPRSV 387
Query: 119 STGLGLSLDNTRLASTGDSALLSLIGDDIDRELQQQDLEMDRFLKLQGEQLRQTILEKVQ 178
STGLGLSLDNT L STGDSALLSLIGDDI RELQQQD E+DRFLK+QGE+LRQ +LEKVQ
Sbjct: 388 STGLGLSLDNTHLTSTGDSALLSLIGDDIKRELQQQDAEIDRFLKVQGERLRQAVLEKVQ 567
Query: 179 ATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMI 238
ATQLQSVS+IEDKVLQKLREKE VE+INKRN+ELEDQMEQL+VEAG+WQQRARYNENMI
Sbjct: 568 ATQLQSVSLIEDKVLQKLREKEAMVESINKRNIELEDQMEQLTVEAGSWQQRARYNENMI 747
Query: 239 AALKFNLQQAYLQGRDSKEGCGDSEVDDTASCCNGRSLDFHLLSNENSNMKDLMKCKACR 298
AALKFNLQQAY+Q RDSKEGCGDSEVDDTASCCNGRSLDFHLLS EN++MK++M CKACR
Sbjct: 748 AALKFNLQQAYVQSRDSKEGCGDSEVDDTASCCNGRSLDFHLLSRENTDMKEMMTCKACR 927
Query: 299 VNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVYM 340
VNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEV M
Sbjct: 928 VNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEV*M 1053
>TC208795 similar to UP|Q9SGY4 (Q9SGY4) F20B24.9, partial (28%)
Length = 799
Score = 150 bits (378), Expect = 1e-36
Identities = 76/182 (41%), Positives = 118/182 (64%), Gaps = 6/182 (3%)
Frame = +2
Query: 165 QGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSVEA 224
Q EQL + + + Q ++ IE + KL+EK+ E+EN+N++N EL ++++Q++VE
Sbjct: 32 QKEQLSKGVRDMKQKHMAALLTSIEKGISTKLKEKDVEIENMNRKNRELAERIKQVAVEV 211
Query: 225 GAWQQRARYNENMIAALKFNLQQAYLQGRD-SKEGCGDSEVDDTASCCNGRSLDFHLLSN 283
+W RA+YNE+++ L+ NLQQA QG + KEG GDSEVDD AS + + L +
Sbjct: 212 QSWHYRAKYNESIVNTLRNNLQQAISQGAEQGKEGFGDSEVDDDASYIDPNNFLNILAAP 391
Query: 284 ENSNMK-----DLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEV 338
NS K + + C+AC+V V+M+L+PC+HLCLCKDCE ++ CP+CQ K +EV
Sbjct: 392 INSTHKSYQDMENLTCRACKVKTVSMLLMPCRHLCLCKDCEGFINVCPICQLIKTASVEV 571
Query: 339 YM 340
++
Sbjct: 572 HL 577
>TC208034 weakly similar to UP|Q9LPJ0 (Q9LPJ0) F6N18.12, partial (46%)
Length = 1183
Score = 139 bits (349), Expect = 2e-33
Identities = 77/213 (36%), Positives = 117/213 (54%)
Frame = +3
Query: 128 NTRLASTGDSALLSLIGDDIDRELQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSI 187
N + S +SL+ + + +++QQ E+D+FL+ Q EQLR+ + EK Q +
Sbjct: 273 NINIRGCHSSPFVSLLSEGLSSQIKQQRDEIDQFLQAQEEQLRRALAEKRQRHYRTLLRA 452
Query: 188 IEDKVLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQ 247
E+ VL++LREKE E+E + N ELE + QLSVEA WQ RA+ E AAL+ LQQ
Sbjct: 453 AEESVLRRLREKEAELEKATRHNAELEARATQLSVEAQLWQARAKAQEATAAALQAQLQQ 632
Query: 248 AYLQGRDSKEGCGDSEVDDTASCCNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLL 307
A + G D + G G SC G + D + + KC+ C ++V+L
Sbjct: 633 AMMIG-DGENGGGGG-----LSCAGGGAEDAESAYVDPDRVGPTPKCRGCAKRVASVVVL 794
Query: 308 PCKHLCLCKDCESKLSFCPLCQSSKFIGMEVYM 340
PC+HLC+C +C++ CP+C + K +EVY+
Sbjct: 795 PCRHLCICAECDTHFRACPVCLTVKNSTVEVYL 893
>TC216836 weakly similar to UP|Q9LPJ0 (Q9LPJ0) F6N18.12, partial (46%)
Length = 1272
Score = 129 bits (324), Expect = 2e-30
Identities = 88/273 (32%), Positives = 134/273 (48%), Gaps = 18/273 (6%)
Frame = +1
Query: 86 FGLEPERKRLKEQDFLENNSQISSVDFLQPRSVSTGLGLSLDNTRLAS------------ 133
F L+ + +L L N+ Q Q VSTGL LS D+
Sbjct: 178 FSLQSQPPQLVHLSQLHNHHQ------QQNNVVSTGLRLSFDDQHFQQQQRLQLHQNESQ 339
Query: 134 ---TGDSALLSLIGDDIDRELQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIED 190
+ SA LSL+ + +++QQ E+D+ L QGEQLR+ + EK Q +S E+
Sbjct: 340 QHRSHSSAFLSLLSQGLGSQIKQQRDEIDQLLHAQGEQLRRALAEKRQRHYRALLSAAEE 519
Query: 191 KVLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYL 250
V ++LREKE EVE ++N ELE + +LSVEA WQ +AR E +L+ LQQ L
Sbjct: 520 AVARQLREKEAEVEMATRKNAELEARAAKLSVEAQVWQAKARAQEATAVSLQTKLQQTIL 699
Query: 251 Q--GRD-SKEGCGDSEVDDTASCCNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLL 307
G D + G + V+ A +D + + KC+ C ++V+L
Sbjct: 700 SHGGEDPAVVGVSSAAVEGQAEDAESAYIDPDRVVAATAARP---KCRGCAKRVASVVVL 870
Query: 308 PCKHLCLCKDCESKLSFCPLCQSSKFIGMEVYM 340
PC+HLC+C +C++ CP+C + K +EV++
Sbjct: 871 PCRHLCVCTECDAHFRACPVCLTPKNSTVEVFL 969
>TC218572 similar to UP|Q9FHE4 (Q9FHE4) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K17O22 (AT5g45100/K17O22_9),
partial (43%)
Length = 716
Score = 102 bits (255), Expect = 2e-22
Identities = 61/197 (30%), Positives = 97/197 (48%), Gaps = 12/197 (6%)
Frame = +1
Query: 146 DIDRELQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVEN 205
D+ Q Q E+DRF+ E++R + E+ ++ I++ V +KL+EK+ E++
Sbjct: 10 DLLFHFQNQQSEIDRFIAQHTEKVRMELEEQRVRQSRMLIAAIQEAVAKKLKEKDEEIQR 189
Query: 206 INKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQ-GRDSKEGCGDSEV 264
+ K N L+++++ L VE W++ A+ NE L+ NL+Q D + V
Sbjct: 190 VGKLNWVLQERVKNLCVENQIWKELAQTNEATANNLRNNLEQVLAHVSEDHHHNLHHTTV 369
Query: 265 DDTASCCNGRSLDFHLLSNE-------NSNMKDLM----KCKACRVNEVTMVLLPCKHLC 313
+ S C + + H E N D + C C V E ++LLPC+HLC
Sbjct: 370 EAAESSCASNNNNSHHREEEEVCGGSGNGKQSDGVLGKRMCNQCGVRESIVLLLPCRHLC 549
Query: 314 LCKDCESKLSFCPLCQS 330
LC CES + CPLCQS
Sbjct: 550 LCTMCESTVRNCPLCQS 600
>TC228566 weakly similar to UP|Q9FHE4 (Q9FHE4) Arabidopsis thaliana genomic
DNA, chromosome 5, TAC clone:K17O22
(AT5g45100/K17O22_9), partial (56%)
Length = 1081
Score = 102 bits (255), Expect = 2e-22
Identities = 79/317 (24%), Positives = 141/317 (43%), Gaps = 15/317 (4%)
Frame = +3
Query: 29 QTIEGQMSQQMAFYNPTDLQDQSQHP-PYIPPFHVVGFAPGPVIPADGSDGGVDLHWNFG 87
Q ++ Q++ + T + S P P P ++ F + +D G+ H
Sbjct: 213 QQLQHQLNSDYMYNTTTQMDSSSALPQPATMPESLLSFYQSNFCDPNKADSGLTYH---- 380
Query: 88 LEPERKRLKEQDFLENNSQISSVDFLQPRSVSTGLGLSLDNTRLASTGDSALLSLIGDDI 147
+ +RKR +DF ++++S+ Q +S+ S + +I
Sbjct: 381 IPLQRKR--SRDF---TTELTSLPAHQKNKISSDP------------------SFLNQEI 491
Query: 148 DRELQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENIN 207
+ Q Q E+DR L E++R + E+ VS I++ + +KL+EK+ E++ +
Sbjct: 492 LYQFQNQQSEIDRVLAHHTEKVRMELEEQKMRQSRMFVSAIQEAMAKKLKEKDQEIQRMG 671
Query: 208 KRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQ--------GRDSKEGC 259
K N L+++++ L +E W++ A+ NE+ L+ NL+Q G D++ C
Sbjct: 672 KLNWTLQERVKSLCMENQIWRELAQTNESTANYLRSNLEQVLAHVGEERATXGDDAQSSC 851
Query: 260 GDSEV----DDTAS--CCNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHLC 313
G ++ +DTA+ GR CK C + E ++LLPC+HLC
Sbjct: 852 GSNDAAEAGNDTAASAAATGRG----------------RLCKNCGLRESVVLLLPCRHLC 983
Query: 314 LCKDCESKLSFCPLCQS 330
LC C S + CP+C S
Sbjct: 984 LCTMCGSTVRNCPICDS 1034
>TC216835 weakly similar to UP|Q9LPJ0 (Q9LPJ0) F6N18.12, partial (47%)
Length = 776
Score = 101 bits (252), Expect = 4e-22
Identities = 60/178 (33%), Positives = 93/178 (51%), Gaps = 2/178 (1%)
Frame = +3
Query: 165 QGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSVEA 224
Q EQL + + EK +S E+ V ++LREKE EVE ++N ELE + +LSVEA
Sbjct: 144 QAEQLXRALAEKXXRHYRALLSTAEEAVARRLREKEAEVEMATRKNAELEARAAKLSVEA 323
Query: 225 GAWQQRARYNENMIAALKFNLQQAYL-QGRDSKEGCG-DSEVDDTASCCNGRSLDFHLLS 282
WQ +AR E A+L+ LQQ + G + G S V+ A +D +
Sbjct: 324 QVWQAKARAQEATAASLQAQLQQTIMSHGGEELAAVGVSSAVEGQAEDAESAYIDPERVV 503
Query: 283 NENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVYM 340
+ KC+ C ++V+LPC+HLC+C +C++ CP+C + K +EV++
Sbjct: 504 VATTARP---KCRGCAKRVASVVVLPCRHLCICTECDAHFRACPVCLTLKNSTVEVFL 668
>TC230232 similar to UP|Q8LAY3 (Q8LAY3) Inhibitor of apoptosis-like protein,
partial (27%)
Length = 939
Score = 89.0 bits (219), Expect = 3e-18
Identities = 62/208 (29%), Positives = 100/208 (47%), Gaps = 9/208 (4%)
Frame = +1
Query: 103 NNSQISSVDFLQPRSVSTGLGLSLDNTRLASTGDSALLSLIGDDIDRELQQQDLEMDRFL 162
NN +S + + R + G + +N DS S +G D+ ++QQQ L+++ +
Sbjct: 322 NNVSVSPMSRKRSRDNNNNYGYNNNNN------DS--FSFLGQDVSLQIQQQQLDIEHLI 477
Query: 163 KLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSV 222
+ E++R I EK + + + IE V++KL+ KE E+E I K N LE++++ L +
Sbjct: 478 MQRMEKVRMEIDEKRKRQARRIIEAIEVGVMKKLKTKEEEIEKIGKLNWALEEKVKHLCM 657
Query: 223 EAGAWQQRARYNENMIAALKFNLQQAYLQ--GRDSKEG-------CGDSEVDDTASCCNG 273
E W+ A NE AL+ NL+Q Q G ++E CG +E+DD SCC
Sbjct: 658 ENQVWRNIAETNEATANALRCNLEQVLAQRGGMAAEEDVGGGATVCGGAEMDDAESCCGS 837
Query: 274 RSLDFHLLSNENSNMKDLMKCKACRVNE 301
D L E + L C + E
Sbjct: 838 TEEDG--LEKETGGWRTLAGCAGVKDKE 915
>BI969751
Length = 734
Score = 88.6 bits (218), Expect = 4e-18
Identities = 55/197 (27%), Positives = 87/197 (43%), Gaps = 13/197 (6%)
Frame = -1
Query: 154 QDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMEL 213
Q E+D ++ E+LR + E+ + + +E L LR+K+ E+ K++ EL
Sbjct: 620 QQREIDXHIRSHNEKLRILLQEQRXXHVAELLKKVESNALHLLRQKDEEIAQATKKSTEL 441
Query: 214 EDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQA-----YLQGRDSKEGCGDSEVDDTA 268
++ M +L VE +W++ A NE M+ +L L+ Y ++ E C D + + A
Sbjct: 440 KEFMTRLEVENQSWRKVAEENEAMVLSLHNTLEDMKERALYRVTKEDAESCCDENMRNRA 261
Query: 269 S--------CCNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCES 320
C G + K M CK C + LPC+HLC CK CE
Sbjct: 260 MEEGTGENRLCGGGGAGG--VEEVEQIRKRTMDCKCCNSQNSCFMFLPCRHLCSCKTCEP 87
Query: 321 KLSFCPLCQSSKFIGME 337
L CP+C K +E
Sbjct: 86 FLQVCPVCSMPKKSSIE 36
>TC230231 similar to UP|Q8LAY3 (Q8LAY3) Inhibitor of apoptosis-like protein,
partial (17%)
Length = 615
Score = 81.3 bits (199), Expect = 6e-16
Identities = 53/168 (31%), Positives = 73/168 (42%), Gaps = 24/168 (14%)
Frame = +2
Query: 197 REKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQ--GRD 254
+ KE E+E I K N + L + W+ A E A++ NL+Q Q G
Sbjct: 2 KTKEEEIEIIGKMNWAXXXXVMHLCMXNQVWRNIAXTKEATANAMRCNLEQVLAQRGGMA 181
Query: 255 SKEG-------CGDSEVDDTASCCNGRSLDFHLLSNENSNMKDLMKC------------- 294
S+E CG +E+DD SCC D L E + L C
Sbjct: 182 SEEDVGVGATVCGGAEMDDAESCCGSTEEDG--LEKETGGWRTLAGCAGVKDKEGGGNGR 355
Query: 295 --KACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVYM 340
+ CR E +++LPC+HLCLC C S L CP+C+S K + V M
Sbjct: 356 LCRNCRKEESCVLILPCRHLCLCTVCGSSLHICPICKSYKTASVHVNM 499
>AW598751
Length = 247
Score = 54.3 bits (129), Expect = 8e-08
Identities = 25/30 (83%), Positives = 26/30 (86%), Gaps = 2/30 (6%)
Frame = +2
Query: 67 PGPVIPADGSDGGVD--LHWNFGLEPERKR 94
PGPV PADGSDGGVD L WN+GLEPERKR
Sbjct: 8 PGPVPPADGSDGGVDLQLQWNYGLEPERKR 97
>TC231131 similar to GB|BAB71852.1|16902294|AB062739 kinesin-related protein
{Arabidopsis thaliana;} , partial (15%)
Length = 725
Score = 52.8 bits (125), Expect = 2e-07
Identities = 51/209 (24%), Positives = 85/209 (40%), Gaps = 28/209 (13%)
Frame = +3
Query: 151 LQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKRN 210
L DL+M+ LQ + R+ LE ++ E + L++ E +VE KR
Sbjct: 45 LDANDLKME----LQARRQREAALE---------AALAEKEFLEE--EYRKKVEEAKKRE 179
Query: 211 MELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQGRDSKEGCGDSEVDDTASC 270
LE+ + A W A+ + A + N+ ++ D + D + +D S
Sbjct: 180 ASLENDL------ANMWVLVAKLKKEGGAVPESNIN---IKKVDEEAHTNDLKTNDIESD 332
Query: 271 CNGRS--LDFHLLSNENSN-----------MKDLMK---------------CKACRVNEV 302
+ LD + NE +N MK++ + CK C +
Sbjct: 333 IIPKEQILDVSIPENEITNEDPLVVRLKARMKEMREKEFKHLGNGDANSHVCKVCFQSST 512
Query: 303 TMVLLPCKHLCLCKDCESKLSFCPLCQSS 331
+LLPC+H CLCK C S CP+C+++
Sbjct: 513 AAILLPCRHFCLCKSCSLACSECPICRTN 599
>TC211501 similar to GB|BAB71852.1|16902294|AB062739 kinesin-related protein
{Arabidopsis thaliana;} , partial (10%)
Length = 775
Score = 50.8 bits (120), Expect = 9e-07
Identities = 18/38 (47%), Positives = 24/38 (62%)
Frame = +3
Query: 294 CKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSS 331
CK C + +LLPC+H CLCK C S CP+C++S
Sbjct: 312 CKVCFESPTAAILLPCRHFCLCKSCSLACSECPICRTS 425
>BE346744 weakly similar to GP|21593126|gb| inhibitor of apoptosis-like
protein {Arabidopsis thaliana}, partial (13%)
Length = 145
Score = 50.4 bits (119), Expect = 1e-06
Identities = 20/36 (55%), Positives = 24/36 (66%)
Frame = -1
Query: 294 CKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQ 329
C C V E ++LLPC+HLCLC C S + CPLCQ
Sbjct: 136 CNQCGVRESIVLLLPCRHLCLCTMCWSTVHNCPLCQ 29
>TC217510
Length = 791
Score = 48.9 bits (115), Expect = 3e-06
Identities = 19/46 (41%), Positives = 26/46 (56%)
Frame = +1
Query: 294 CKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVY 339
C+ C E+ +VLLPC+H LC C K CP+C+ S + VY
Sbjct: 334 CRVCFEGEINVVLLPCRHRVLCSTCSEKCKKCPICRDSIAERLPVY 471
>CO985810
Length = 910
Score = 48.1 bits (113), Expect = 6e-06
Identities = 49/193 (25%), Positives = 87/193 (44%), Gaps = 11/193 (5%)
Frame = -3
Query: 149 RELQQ--QDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDK--VLQKLREKETEVE 204
R+L Q Q+LE R + Q E Q + + LQ+ SI +++ + + + KE ++
Sbjct: 692 RKLAQLLQELEQARMQQEQVEGRWQQEAKAKEEFILQASSIKKEREQIEESGKSKEDAIK 513
Query: 205 NINKRNMEL-EDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQG-RDSKEGCGDS 262
+RN ++ D + +L E Q R + + + IAAL+ + Y D K G
Sbjct: 512 LKAERNRQMYRDDIHKLEKEIS--QLRLKTDSSKIAALRMGIDGCYASKCLDMKNGTAQK 339
Query: 263 EVDDTASCCNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDC---- 318
E AS + +D + +K +C C E++++ +PC H +CK C
Sbjct: 338 E--PRASFISELVIDH----SATGGVKREQECVMCLSEEMSVLFMPCAHQVVCKTCNELH 177
Query: 319 -ESKLSFCPLCQS 330
+ + CP C+S
Sbjct: 176 EKQGMQDCPSCRS 138
>TC208035 similar to UP|Q9LPJ0 (Q9LPJ0) F6N18.12, partial (15%)
Length = 445
Score = 47.0 bits (110), Expect = 1e-05
Identities = 16/44 (36%), Positives = 28/44 (63%)
Frame = +2
Query: 297 CRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVYM 340
C ++V+LPC+HLC+C +C+ CP+C + K ++VY+
Sbjct: 44 CAKRVASVVVLPCRHLCICAECDGHFRACPVCLTVKNSTIQVYL 175
>CD398985 similar to PIR|T01393|T013 apoptosis inhibitor homolog T4I9.12 -
Arabidopsis thaliana, partial (9%)
Length = 697
Score = 45.4 bits (106), Expect = 4e-05
Identities = 37/157 (23%), Positives = 69/157 (43%), Gaps = 7/157 (4%)
Frame = -1
Query: 181 QLQSVSIIEDKVLQKLREKETEVENINKRNMEL-EDQMEQLSVEAGAWQQRARYNENMIA 239
Q S+ +++ + + KE ++ +RN ++ D + +L E Q R + + + IA
Sbjct: 607 QASSIKKEREQIEESGKSKEDAIKLKAERNRQMYRDDIHKLEKEIS--QLRLKTDSSKIA 434
Query: 240 ALKFNLQQAYLQG-RDSKEGCGDSEVDDTASCCNGRSLDFHLLSNENSNMKDLMKCKACR 298
AL+ + Y D K G E AS + +D + +K +C C
Sbjct: 433 ALRMGIDGCYASKCLDMKNGTAQKE--PRASFISELVIDH----SATGGVKREQECVMCL 272
Query: 299 VNEVTMVLLPCKHLCLCKDC-----ESKLSFCPLCQS 330
E++++ +PC H +CK C + + CP C+S
Sbjct: 271 SEEMSVLFMPCAHQVVCKTCNELHEKQGMQDCPSCRS 161
>BM523427
Length = 419
Score = 44.3 bits (103), Expect = 8e-05
Identities = 19/42 (45%), Positives = 23/42 (54%)
Frame = +1
Query: 288 MKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQ 329
M DL C C E V +PC H+C C C S L+ CPLC+
Sbjct: 61 MPDL--CVICLEQEYNAVFVPCGHMCCCTTCSSHLTNCPLCR 180
>TC208697
Length = 769
Score = 44.3 bits (103), Expect = 8e-05
Identities = 28/85 (32%), Positives = 38/85 (43%)
Frame = +3
Query: 245 LQQAYLQGRDSKEGCGDSEVDDTASCCNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTM 304
LQ+ L K G +++V+ S +G D M DL C C E
Sbjct: 105 LQRRVLAAAAKKSG-QNNDVEKADSLSDGAKKD--------RLMPDL--CVICLEQEYNA 251
Query: 305 VLLPCKHLCLCKDCESKLSFCPLCQ 329
V +PC H+C C C S L+ CPLC+
Sbjct: 252 VFVPCGHMCCCTACSSHLTNCPLCR 326
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,993,922
Number of Sequences: 63676
Number of extensions: 203689
Number of successful extensions: 1548
Number of sequences better than 10.0: 108
Number of HSP's better than 10.0 without gapping: 1483
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1521
length of query: 340
length of database: 12,639,632
effective HSP length: 98
effective length of query: 242
effective length of database: 6,399,384
effective search space: 1548650928
effective search space used: 1548650928
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148486.4


