

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148486.10 - phase: 0
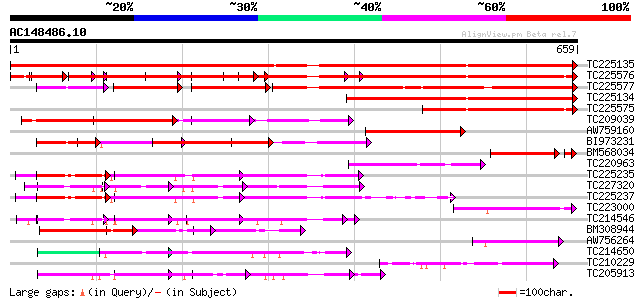
(659 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC225135 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, ... 982 0.0
TC225576 similar to UP|Q9AT32 (Q9AT32) Poly(A)-binding protein, ... 620 e-178
TC225577 similar to UP|Q9M6E4 (Q9M6E4) Poly(A)-binding protein (... 473 e-134
TC225134 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, ... 397 e-111
TC225575 similar to UP|Q9M6E4 (Q9M6E4) Poly(A)-binding protein (... 295 4e-80
TC209039 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, ... 272 3e-73
AW759160 weakly similar to GP|7528270|gb|A poly(A)-binding prote... 134 1e-31
BI973231 129 3e-30
BM568034 similar to GP|7673359|gb|A poly(A)-binding protein {Nic... 117 3e-28
TC220963 similar to GB|AAA32832.1|166786|ATHPABP poly(A)-binding... 118 7e-27
TC225235 similar to GB|AAP37853.1|30725662|BT008494 At1g11650 {A... 107 1e-23
TC227320 similar to UP|Q93YF1 (Q93YF1) Nucleic acid binding prot... 107 2e-23
TC225237 similar to UP|Q9LEB4 (Q9LEB4) RNA Binding Protein 45, p... 103 2e-22
TC223000 similar to GB|AAG02117.1|9930616|AF293840 poly(A) bindi... 103 3e-22
TC214546 similar to PIR|T01932|T01932 RNA binding protein homolo... 95 9e-20
BM308944 92 7e-19
AW756264 weakly similar to GP|13560783|gb| poly(A)-binding prote... 90 4e-18
TC214650 similar to UP|Q9LJH8 (Q9LJH8) RNA binding protein nucle... 89 5e-18
TC210229 similar to UP|O57336 (O57336) Polyadenylate binding pro... 87 3e-17
TC205913 weakly similar to UP|Q9LEB4 (Q9LEB4) RNA Binding Protei... 87 3e-17
>TC225135 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, partial
(97%)
Length = 2392
Score = 982 bits (2539), Expect = 0.0
Identities = 496/663 (74%), Positives = 560/663 (83%), Gaps = 4/663 (0%)
Frame = +3
Query: 1 MAQIQVQHQTPAPVPAPSNGVVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVS 60
MAQ+QV Q P P G NQFVTTSLYVGDL+ NV D+QLYDLFNQ+GQVVS
Sbjct: 90 MAQVQVPPQNAMPGPNGGGGA-GGAGNQFVTTSLYVGDLDPNVTDAQLYDLFNQLGQVVS 266
Query: 61 VRVCRDLATRRSLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPSSRKSGTA 120
VRVCRDL +RRSLGYGYVNF+NPQDAARALDVLNFTP+NN+ IR+MYSHRDPS RKSG
Sbjct: 267 VRVCRDLTSRRSLGYGYVNFSNPQDAARALDVLNFTPLNNRPIRIMYSHRDPSIRKSGQG 446
Query: 121 NIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKL 180
NIFIKNLD+ IDHKALHDTFS+FG I+SCK+ATD SGQSKGYGFVQF+ E+SAQ AI+KL
Sbjct: 447 NIFIKNLDRAIDHKALHDTFSTFGNILSCKVATDSSGQSKGYGFVQFDNEESAQKAIEKL 626
Query: 181 NGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGAYGTITSA 240
NGML+NDKQV+VG FLRKQ+R++ K KFNNV+VKNLSES T+D+LKN FG +GTITSA
Sbjct: 627 NGMLLNDKQVYVGPFLRKQERESAADKAKFNNVFVKNLSESTTDDELKNTFGEFGTITSA 806
Query: 241 VLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSEREQELKGRF 300
V+MRD DG+SKCFGFVNFENA+DAA+AVEALNGKK DD+EWYVGKAQKKSERE ELK RF
Sbjct: 807 VVMRDGDGKSKCFGFVNFENADDAARAVEALNGKKFDDKEWYVGKAQKKSERENELKQRF 986
Query: 301 EQTVKESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMR 360
EQ++KE+ DK+QG NLY+KNLDDSI DEKLKE+FS FGTITS KV MR
Sbjct: 987 EQSMKEA-ADKYQGANLYVKNLDDSIGDEKLKELFSPFGTITSCKV------------MR 1127
Query: 361 DPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARLQAQFSQMR 420
DPNG+SRGSGFVAFSTPEEASRAL EMNGKM+VSKPLYV +AQRKEDRRARLQAQF+QMR
Sbjct: 1128DPNGLSRGSGFVAFSTPEEASRALLEMNGKMVVSKPLYVTLAQRKEDRRARLQAQFAQMR 1307
Query: 421 PVAITPSVAPRMPLYPPGTPGLGQQFMYGQGPPAMMPPQAGFGYQQQLVPGMRPGGGPMP 480
PV + PSV PR+P+YPPG PG+GQQ Y QGPPA++P Q GFGYQQQL+PGMRPG P+P
Sbjct: 1308PVGMPPSVGPRVPMYPPGGPGIGQQLFYSQGPPAIIPSQPGFGYQQQLMPGMRPGAAPVP 1487
Query: 481 SYFVPMVQQGQQGQRPGGRRGGPGQQPPQQVPMMQQQMLPRQRVYRYPPGRSNIQDAPVQ 540
++FVPMVQQGQQGQRPGGRR G QQ Q VPMM QQMLPR RVYRYPPGR I D P+
Sbjct: 1488NFFVPMVQQGQQGQRPGGRRPGAVQQSQQPVPMMPQQMLPRGRVYRYPPGR-GIPDVPIP 1664
Query: 541 NIGGGMMS--YDMGGLPLRD--VVPPMPIHALATALANAPPEQQRTMLGEALYPLVDQLE 596
+ GGM S YD+GG+PLRD + +P+ ALATALANA PEQQRTMLGE LYPLV+QLE
Sbjct: 1665GVAGGMFSVPYDVGGMPLRDASISQQIPVGALATALANASPEQQRTMLGENLYPLVEQLE 1844
Query: 597 HDSAAKVTGMLLEMDQPEVLHLIESPDALKAKVAEAMEVLRNVSQQQGNSPADQLASLSL 656
D+AAKVTGMLLEMDQ EVLHL+ESP+ALKAKVAEAM+VLRNV+QQQ ADQLASLSL
Sbjct: 1845PDNAAKVTGMLLEMDQTEVLHLLESPEALKAKVAEAMDVLRNVAQQQAGGTADQLASLSL 2024
Query: 657 NDS 659
ND+
Sbjct: 2025NDN 2033
>TC225576 similar to UP|Q9AT32 (Q9AT32) Poly(A)-binding protein, partial (93%)
Length = 2491
Score = 620 bits (1600), Expect = e-178
Identities = 324/398 (81%), Positives = 349/398 (87%), Gaps = 4/398 (1%)
Frame = +1
Query: 266 KAVEALNGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDS 325
K ++ L G+K+ VGKAQKKSEREQELKGRFEQ++KES DK+QG+NLYLKNLDD+
Sbjct: 1063 KLLKDLMGRKLMIRSGXVGKAQKKSEREQELKGRFEQSIKESA-DKYQGVNLYLKNLDDT 1239
Query: 326 ITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALG 385
I+DEKLKEMF+E+GTITS KV MRDP G+ RGSGFVAFSTPEEASRALG
Sbjct: 1240 ISDEKLKEMFAEYGTITSCKV------------MRDPTGIGRGSGFVAFSTPEEASRALG 1383
Query: 386 EMNGKMIVSKPLYVAVAQRKEDRRARLQAQFSQMRPVAITPSVAPRMPLYPPGTPGLGQQ 445
EMNGKM KPLYVA+AQRKE+RRARLQAQFSQMRPVAITPSVAPRMPLYPPG PGLGQQ
Sbjct: 1384 EMNGKMFAGKPLYVALAQRKEERRARLQAQFSQMRPVAITPSVAPRMPLYPPGAPGLGQQ 1563
Query: 446 FMYGQGPPAMMPPQAGFGYQQQLVPGMRPGGGPMPSYFVPMVQQGQQGQRPGGRRG-GPG 504
F+YGQGPPAMMPPQAGFGYQQQLVPGMRPGGGPMPS+FVPMVQQGQQGQRP GRRG GP
Sbjct: 1564 FLYGQGPPAMMPPQAGFGYQQQLVPGMRPGGGPMPSFFVPMVQQGQQGQRPVGRRGTGPV 1743
Query: 505 QQPPQQVPMMQQQMLPRQRVYRYPPGRSNIQDAPVQNIGGGMMS--YDMGGLPLRDVV-P 561
QQP Q +PMMQQQMLPR RVYRYPPGR N+QD P+Q GGM+S YDMGGLP+RD V
Sbjct: 1744 QQPQQPMPMMQQQMLPRGRVYRYPPGR-NMQDVPLQVAAGGMLSVPYDMGGLPMRDAVGQ 1920
Query: 562 PMPIHALATALANAPPEQQRTMLGEALYPLVDQLEHDSAAKVTGMLLEMDQPEVLHLIES 621
PMPI ALATALANAPPEQQRTMLGEALYPLVDQLEHDSAAKVTGMLLEMDQPEVLHLIES
Sbjct: 1921 PMPIQALATALANAPPEQQRTMLGEALYPLVDQLEHDSAAKVTGMLLEMDQPEVLHLIES 2100
Query: 622 PDALKAKVAEAMEVLRNVSQQQGNSPADQLASLSLNDS 659
PDALKAKVAEAM+VLRNV+QQQ N PADQLASLSLND+
Sbjct: 2101 PDALKAKVAEAMDVLRNVAQQQTN-PADQLASLSLNDN 2211
Score = 377 bits (969), Expect(2) = e-129
Identities = 182/221 (82%), Positives = 202/221 (91%)
Frame = +3
Query: 69 TRRSLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPSSRKSGTANIFIKNLD 128
TRRSLGYGYVNF+NPQDAARALDVLNFTP+NN+ IR+MYSHRDPS RKSGTANIFIKNLD
Sbjct: 471 TRRSLGYGYVNFSNPQDAARALDVLNFTPLNNRPIRIMYSHRDPSLRKSGTANIFIKNLD 650
Query: 129 KTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDK 188
K IDHKALHDTFSSFG I+SCKIATD SG SKGYGFVQF+ E++AQNAIDKLNGMLINDK
Sbjct: 651 KAIDHKALHDTFSSFGLILSCKIATDASGLSKGYGFVQFDNEEAAQNAIDKLNGMLINDK 830
Query: 189 QVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADG 248
QV+VGHFLRKQDR+N LSKTKFNNVYVKNLSES T+++L FG YGTITSAV+MRDADG
Sbjct: 831 QVYVGHFLRKQDRENALSKTKFNNVYVKNLSESTTDEELMKFFGEYGTITSAVIMRDADG 1010
Query: 249 RSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKK 289
+S+CFGFVNFEN +DAAKAVE LNGKKVDD+EW K+ +K
Sbjct: 1011KSRCFGFVNFENPDDAAKAVEGLNGKKVDDKEWXCWKSPEK 1133
Score = 145 bits (366), Expect = 6e-35
Identities = 86/238 (36%), Positives = 130/238 (54%)
Frame = +3
Query: 158 QSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKN 217
+S GYG+V F A A+D LN +N++ + + + RD L K+ N+++KN
Sbjct: 477 RSLGYGYVNFSNPQDAARALDVLNFTPLNNRPIRIMY----SHRDPSLRKSGTANIFIKN 644
Query: 218 LSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKVD 277
L ++ L + F ++G I S + DA G SK +GFV F+N E A A++ LNG ++
Sbjct: 645 LDKAIDHKALHDTFSSFGLILSCKIATDASGLSKGYGFVQFDNEEAAQNAIDKLNGMLIN 824
Query: 278 DEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKLKEMFSE 337
D++ YVG +K +RE L K + N+Y+KNL +S TDE+L + F E
Sbjct: 825 DKQVYVGHFLRKQDRENAL-------------SKTKFNNVYVKNLSESTTDEELMKFFGE 965
Query: 338 FGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSK 395
+GTITS VIMRD +G SR GFV F P++A++A+ +NGK + K
Sbjct: 966 YGTITS------------AVIMRDADGKSRCFGFVNFENPDDAAKAVEGLNGKKVDDK 1103
Score = 102 bits (255), Expect(2) = e-129
Identities = 55/69 (79%), Positives = 57/69 (81%), Gaps = 2/69 (2%)
Frame = +1
Query: 1 MAQIQVQHQTPAPVPAPSNGVV--PNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQV 58
MAQIQVQHQ+P P P NGV PN NQFVTTSLYVGDLE NVND+QLYDLFNQV QV
Sbjct: 265 MAQIQVQHQSPVSAPPP-NGVANAPNNPNQFVTTSLYVGDLEQNVNDAQLYDLFNQVVQV 441
Query: 59 VSVRVCRDL 67
VSVRVCRDL
Sbjct: 442 VSVRVCRDL 468
Score = 82.0 bits (201), Expect = 8e-16
Identities = 54/164 (32%), Positives = 82/164 (49%)
Frame = +3
Query: 249 RSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESV 308
RS +G+VNF N +DAA+A++ LN +++ + S R+ L+
Sbjct: 477 RSLGYGYVNFSNPQDAARALDVLNFTPLNNRPIRI----MYSHRDPSLR----------- 611
Query: 309 VDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRG 368
K N+++KNLD +I + L + FS FG I S K+ T D +G+S+G
Sbjct: 612 --KSGTANIFIKNLDKAIDHKALHDTFSSFGLILSCKIAT------------DASGLSKG 749
Query: 369 SGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARL 412
GFV F E A A+ ++NG +I K +YV RK+DR L
Sbjct: 750 YGFVQFDNEEAAQNAIDKLNGMLINDKQVYVGHFLRKQDRENAL 881
Score = 78.2 bits (191), Expect = 1e-14
Identities = 38/91 (41%), Positives = 56/91 (60%)
Frame = +1
Query: 212 NVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEAL 271
N+Y+KNL ++ +++ LK F YGTITS +MRD G + GFV F E+A++A+ +
Sbjct: 1210 NLYLKNLDDTISDEKLKEMFAEYGTITSCKVMRDPTGIGRGSGFVAFSTPEEASRALGEM 1389
Query: 272 NGKKVDDEEWYVGKAQKKSEREQELKGRFEQ 302
NGK + YV AQ+K ER L+ +F Q
Sbjct: 1390 NGKMFAGKPLYVALAQRKEERRARLQAQFSQ 1482
Score = 68.6 bits (166), Expect(2) = 1e-20
Identities = 32/89 (35%), Positives = 52/89 (57%)
Frame = +1
Query: 113 SSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDS 172
S+ K N+++KNLD TI + L + F+ +G I SCK+ D +G +G GFV F +
Sbjct: 1186 SADKYQGVNLYLKNLDDTISDEKLKEMFAEYGTITSCKVMRDPTGIGRGSGFVAFSTPEE 1365
Query: 173 AQNAIDKLNGMLINDKQVFVGHFLRKQDR 201
A A+ ++NG + K ++V RK++R
Sbjct: 1366 ASRALGEMNGKMFAGKPLYVALAQRKEER 1452
Score = 50.1 bits (118), Expect(2) = 1e-20
Identities = 26/78 (33%), Positives = 45/78 (57%)
Frame = +3
Query: 24 NVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNP 83
N ++ ++YV +L + D +L F + G + S + RD A +S +G+VNF NP
Sbjct: 873 NALSKTKFNNVYVKNLSESTTDEELMKFFGEYGTITSAVIMRD-ADGKSRCFGFVNFENP 1049
Query: 84 QDAARALDVLNFTPMNNK 101
DAA+A++ LN +++K
Sbjct: 1050DDAAKAVEGLNGKKVDDK 1103
Score = 49.3 bits (116), Expect = 5e-06
Identities = 27/90 (30%), Positives = 50/90 (55%)
Frame = +1
Query: 26 ANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQD 85
A+++ +LY+ +L+ ++D +L ++F + G + S +V RD T G G+V F+ P++
Sbjct: 1189 ADKYQGVNLYLKNLDDTISDEKLKEMFAEYGTITSCKVMRD-PTGIGRGSGFVAFSTPEE 1365
Query: 86 AARALDVLNFTPMNNKSIRVMYSHRDPSSR 115
A+RAL +N K + V + R R
Sbjct: 1366 ASRALGEMNGKMFAGKPLYVALAQRKEERR 1455
>TC225577 similar to UP|Q9M6E4 (Q9M6E4) Poly(A)-binding protein (Fragment),
partial (68%)
Length = 1180
Score = 473 bits (1218), Expect = e-134
Identities = 247/355 (69%), Positives = 283/355 (79%), Gaps = 1/355 (0%)
Frame = +3
Query: 306 ESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGV 365
+ VDK+ G NLY+KNLDDS+ DE+L E+FSEFGTITS KV MRDPNG+
Sbjct: 3 KETVDKYHGTNLYIKNLDDSVGDEELMELFSEFGTITSCKV------------MRDPNGI 146
Query: 366 SRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARLQAQFSQMRPVAIT 425
SRGSGFV+FS E A+RALGEMNGKM+ KPLYVA+AQRKEDRRARLQAQFSQ RP AIT
Sbjct: 147 SRGSGFVSFSIAEGATRALGEMNGKMVAGKPLYVALAQRKEDRRARLQAQFSQSRPAAIT 326
Query: 426 PSVAPRMPLYPPGTPGLGQQFMYGQGPPAMMPPQAGFGYQQQLVPGMRPGGGPMPSYFVP 485
P+V+PRMPLYP G P +GQQ +YGQ P +P QA FGYQQ VPGMRPGG P +++VP
Sbjct: 327 PNVSPRMPLYPLGAPAIGQQLLYGQAAPTTIP-QAAFGYQQHFVPGMRPGGAP--NFYVP 497
Query: 486 MVQQGQQGQRPGGRRG-GPGQQPPQQVPMMQQQMLPRQRVYRYPPGRSNIQDAPVQNIGG 544
+VQQGQQGQR GG+RG GP QQ Q +P+M QQMLPR VYRYPPG SN+Q+ + + G
Sbjct: 498 LVQQGQQGQRSGGQRGAGPMQQLQQPLPLMHQQMLPRGHVYRYPPG-SNMQNIQLAGVPG 674
Query: 545 GMMSYDMGGLPLRDVVPPMPIHALATALANAPPEQQRTMLGEALYPLVDQLEHDSAAKVT 604
GM+SY +G PMP ALA+ALANA PEQQRTMLGEALYPLVD+LEH++AAKVT
Sbjct: 675 GMVSYGIG--------QPMPTQALASALANATPEQQRTMLGEALYPLVDKLEHETAAKVT 830
Query: 605 GMLLEMDQPEVLHLIESPDALKAKVAEAMEVLRNVSQQQGNSPADQLASLSLNDS 659
GMLLEMDQPEVLHLIESPDALKAKV EAM+VLR V+QQQ NSPADQLASLSLND+
Sbjct: 831 GMLLEMDQPEVLHLIESPDALKAKVVEAMDVLRKVTQQQSNSPADQLASLSLNDN 995
Score = 79.3 bits (194), Expect = 5e-15
Identities = 39/92 (42%), Positives = 59/92 (63%)
Frame = +3
Query: 212 NVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEAL 271
N+Y+KNL +S +++L F +GTITS +MRD +G S+ GFV+F AE A +A+ +
Sbjct: 33 NLYIKNLDDSVGDEELMELFSEFGTITSCKVMRDPNGISRGSGFVSFSIAEGATRALGEM 212
Query: 272 NGKKVDDEEWYVGKAQKKSEREQELKGRFEQT 303
NGK V + YV AQ+K +R L+ +F Q+
Sbjct: 213 NGKMVAGKPLYVALAQRKEDRRARLQAQFSQS 308
Score = 72.0 bits (175), Expect = 8e-13
Identities = 33/81 (40%), Positives = 51/81 (62%)
Frame = +3
Query: 121 NIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKL 180
N++IKNLD ++ + L + FS FG I SCK+ D +G S+G GFV F + A A+ ++
Sbjct: 33 NLYIKNLDDSVGDEELMELFSEFGTITSCKVMRDPNGISRGSGFVSFSIAEGATRALGEM 212
Query: 181 NGMLINDKQVFVGHFLRKQDR 201
NG ++ K ++V RK+DR
Sbjct: 213 NGKMVAGKPLYVALAQRKEDR 275
Score = 48.5 bits (114), Expect = 9e-06
Identities = 28/84 (33%), Positives = 47/84 (55%)
Frame = +3
Query: 32 TSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALD 91
T+LY+ +L+ +V D +L +LF++ G + S +V RD S G G+V+F+ + A RAL
Sbjct: 30 TNLYIKNLDDSVGDEELMELFSEFGTITSCKVMRD-PNGISRGSGFVSFSIAEGATRALG 206
Query: 92 VLNFTPMNNKSIRVMYSHRDPSSR 115
+N + K + V + R R
Sbjct: 207 EMNGKMVAGKPLYVALAQRKEDRR 278
>TC225134 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, partial
(42%)
Length = 1090
Score = 397 bits (1020), Expect = e-111
Identities = 200/272 (73%), Positives = 227/272 (82%), Gaps = 4/272 (1%)
Frame = +2
Query: 392 IVSKPLYVAVAQRKEDRRARLQAQFSQMRPVAITPSVAPRMPLYPPGTPGLGQQFMYGQG 451
+VSKPLYV +AQRKEDRRARLQAQF+QMRPV + PSV PR+P+YPPG PG+GQQ Y QG
Sbjct: 2 VVSKPLYVTLAQRKEDRRARLQAQFAQMRPVGMPPSVGPRVPMYPPGGPGIGQQIFYAQG 181
Query: 452 PPAMMPPQAGFGYQQQLVPGMRPGGGPMPSYFVPMVQQGQQGQRPGGRRGGPGQQPPQQV 511
PPA++P Q GFGYQQQLVPGMRPG P+P++FVPMVQQGQQGQRPGGRR G QQ Q V
Sbjct: 182 PPAIIPSQPGFGYQQQLVPGMRPGAAPVPNFFVPMVQQGQQGQRPGGRRTGAVQQSQQPV 361
Query: 512 PMMQQQMLPRQRVYRYPPGRSNIQDAPVQNIGGGMMS--YDMGGLPLRD--VVPPMPIHA 567
PMM QQMLPR RVYRYPPGR + D + + GGM S YD+GG+PLRD + +P+ A
Sbjct: 362 PMMPQQMLPRGRVYRYPPGR-GMPDVSMPGVAGGMFSVPYDVGGMPLRDASISQQIPVGA 538
Query: 568 LATALANAPPEQQRTMLGEALYPLVDQLEHDSAAKVTGMLLEMDQPEVLHLIESPDALKA 627
LATALANA PEQQRTMLGE LYPLV+QLE D+AAKVTGMLLEMDQ EVLHL+ESP+ALKA
Sbjct: 539 LATALANASPEQQRTMLGENLYPLVEQLEPDNAAKVTGMLLEMDQTEVLHLLESPEALKA 718
Query: 628 KVAEAMEVLRNVSQQQGNSPADQLASLSLNDS 659
KVAEAM+VLRNV+QQQ ADQLASLSLND+
Sbjct: 719 KVAEAMDVLRNVAQQQAGGTADQLASLSLNDN 814
>TC225575 similar to UP|Q9M6E4 (Q9M6E4) Poly(A)-binding protein (Fragment),
partial (38%)
Length = 791
Score = 295 bits (755), Expect = 4e-80
Identities = 156/184 (84%), Positives = 166/184 (89%), Gaps = 4/184 (2%)
Frame = +2
Query: 480 PSYFVPMVQQGQQGQRPGGRRG-GPGQQPPQQVPMMQQQMLPRQRVYRYPPGRSNIQDAP 538
PS+FVPMVQQ QQGQRPGGRRG GP QQP Q +PMMQQQMLPR RVYRYPPGR N+QD P
Sbjct: 2 PSFFVPMVQQDQQGQRPGGRRGTGPVQQPQQPMPMMQQQMLPRGRVYRYPPGR-NMQDVP 178
Query: 539 VQNIGGGMMS--YDMGGLPLRDVV-PPMPIHALATALANAPPEQQRTMLGEALYPLVDQL 595
+Q + GGMMS YDMGGLP+RD V PMPI ALATALANAPPEQQRTMLGEALYPLVDQL
Sbjct: 179 LQGVAGGMMSVPYDMGGLPIRDAVGQPMPIQALATALANAPPEQQRTMLGEALYPLVDQL 358
Query: 596 EHDSAAKVTGMLLEMDQPEVLHLIESPDALKAKVAEAMEVLRNVSQQQGNSPADQLASLS 655
EHD+AAKVTGMLLEMDQPEVLHLIESPDALKAKVAEAM+VLRNV+QQQ N PADQLASLS
Sbjct: 359 EHDAAAKVTGMLLEMDQPEVLHLIESPDALKAKVAEAMDVLRNVAQQQTN-PADQLASLS 535
Query: 656 LNDS 659
LND+
Sbjct: 536 LNDN 547
>TC209039 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, partial
(25%)
Length = 800
Score = 272 bits (696), Expect = 3e-73
Identities = 133/182 (73%), Positives = 157/182 (86%)
Frame = +1
Query: 14 VPAPSNGVVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSL 73
+ A ++G P++ T SLYVGDL +VND QLYDLFNQV QVVSVR+CRD+AT++SL
Sbjct: 265 IDAAASGANPSLT----TISLYVGDLHHDVNDPQLYDLFNQVAQVVSVRICRDVATQQSL 432
Query: 74 GYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPSSRKSGTANIFIKNLDKTIDH 133
GYGYVNF+N DAA+A+DVLNFTP+N K IR+MYS RDPS+RKSG AN+FIKNLDK IDH
Sbjct: 433 GYGYVNFSNAHDAAKAIDVLNFTPLNGKIIRIMYSIRDPSARKSGAANVFIKNLDKAIDH 612
Query: 134 KALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVG 193
KAL+DTFS+FG I+SCK+ATD SGQSKG+GFVQFE+E+SAQNAIDKLNGMLINDK VFVG
Sbjct: 613 KALYDTFSAFGNILSCKVATDASGQSKGHGFVQFESEESAQNAIDKLNGMLINDKXVFVG 792
Query: 194 HF 195
F
Sbjct: 793 PF 798
Score = 100 bits (250), Expect = 2e-21
Identities = 62/188 (32%), Positives = 100/188 (52%), Gaps = 1/188 (0%)
Frame = +1
Query: 98 MNNKSIRVMYSHRDPSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATD-GS 156
+ N +I S +PS T ++++ +L ++ L+D F+ Q++S +I D +
Sbjct: 250 LENPTIDAAASGANPSLT---TISLYVGDLHHDVNDPQLYDLFNQVAQVVSVRICRDVAT 420
Query: 157 GQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVK 216
QS GYG+V F A AID LN +N K + + + +R D K+ NV++K
Sbjct: 421 QQSLGYGYVNFSNAHDAAKAIDVLNFTPLNGKIIRIMYSIR----DPSARKSGAANVFIK 588
Query: 217 NLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKV 276
NL ++ L + F A+G I S + DA G+SK GFV FE+ E A A++ LNG +
Sbjct: 589 NLDKAIDHKALYDTFSAFGNILSCKVATDASGQSKGHGFVQFESEESAQNAIDKLNGMLI 768
Query: 277 DDEEWYVG 284
+D+ +VG
Sbjct: 769 NDKXVFVG 792
Score = 84.7 bits (208), Expect = 1e-16
Identities = 59/189 (31%), Positives = 92/189 (48%), Gaps = 1/189 (0%)
Frame = +1
Query: 212 NVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRD-ADGRSKCFGFVNFENAEDAAKAVEA 270
++YV +L + L + F + S + RD A +S +G+VNF NA DAAKA++
Sbjct: 310 SLYVGDLHHDVNDPQLYDLFNQVAQVVSVRICRDVATQQSLGYGYVNFSNAHDAAKAIDV 489
Query: 271 LNGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEK 330
LN ++ GK R ++++ K N+++KNLD +I +
Sbjct: 490 LNFTPLN------GKII-----------RIMYSIRDPSARKSGAANVFIKNLDKAIDHKA 618
Query: 331 LKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGK 390
L + FS FG I S KV T D +G S+G GFV F + E A A+ ++NG
Sbjct: 619 LYDTFSAFGNILSCKVAT------------DASGQSKGHGFVQFESEESAQNAIDKLNGM 762
Query: 391 MIVSKPLYV 399
+I K ++V
Sbjct: 763 LINDKXVFV 789
>AW759160 weakly similar to GP|7528270|gb|A poly(A)-binding protein {Cucumis
sativus}, partial (15%)
Length = 352
Score = 134 bits (338), Expect = 1e-31
Identities = 62/116 (53%), Positives = 79/116 (67%)
Frame = +2
Query: 414 AQFSQMRPVAITPSVAPRMPLYPPGTPGLGQQFMYGQGPPAMMPPQAGFGYQQQLVPGMR 473
AQF+QMRPV + PSV+P MP+YPP P + QQ Y Q PP ++P Q F YQQQL+P ++
Sbjct: 2 AQFAQMRPVGMPPSVSPHMPMYPPNGPDINQQIFYTQNPPTIIPSQPRFKYQQQLIPNIK 181
Query: 474 PGGGPMPSYFVPMVQQGQQGQRPGGRRGGPGQQPPQQVPMMQQQMLPRQRVYRYPP 529
PMP++F P+VQQ QQ QRPGG QQ Q VP+M QQ+L + +YRYPP
Sbjct: 182 RNTAPMPNFFRPIVQQKQQKQRPGGEHTDAVQQCQQPVPIMPQQILT*KHIYRYPP 349
>BI973231
Length = 422
Score = 129 bits (325), Expect = 3e-30
Identities = 64/140 (45%), Positives = 96/140 (67%)
Frame = +2
Query: 167 FEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDD 226
FE+E+S+ AI+KLNG + DKQ++VG F++K DR ++ N+Y+KNL +E
Sbjct: 2 FESEESSNVAIEKLNGSTVGDKQLYVGKFVKKSDRILPGPDARYTNLYMKNLDLDVSEAT 181
Query: 227 LKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKA 286
L+ +F ++G I S V+ +D G SK FGFVN++N +DA +A+EA+NG K+ + YV +A
Sbjct: 182 LQEKFSSFGKIVSLVIAKDNIGMSKGFGFVNYDNPDDAKRAMEAMNGSKLGSKILYVARA 361
Query: 287 QKKSEREQELKGRFEQTVKE 306
QKK+EREQ L +FE+ KE
Sbjct: 362 QKKAEREQILHHQFEEKRKE 421
Score = 99.8 bits (247), Expect = 4e-21
Identities = 65/163 (39%), Positives = 88/163 (53%)
Frame = +2
Query: 258 FENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNL 317
FE+ E + A+E LNG V D++ YVGK KKS+R L G + NL
Sbjct: 2 FESEESSNVAIEKLNGSTVGDKQLYVGKFVKKSDRI--LPGPDARYT-----------NL 142
Query: 318 YLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTP 377
Y+KNLD +++ L+E FS FG I S VI +D G+S+G GFV + P
Sbjct: 143 YMKNLDLDVSEATLQEKFSSFGKIVSL------------VIAKDNIGMSKGFGFVNYDNP 286
Query: 378 EEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARLQAQFSQMR 420
++A RA+ MNG + SK LYVA AQ+K +R L QF + R
Sbjct: 287 DDAKRAMEAMNGSKLGSKILYVARAQKKAEREQILHHQFEEKR 415
Score = 75.9 bits (185), Expect = 5e-14
Identities = 41/130 (31%), Positives = 72/130 (54%), Gaps = 4/130 (3%)
Frame = +2
Query: 80 FTNPQDAARALDVLNFTPMNNKSIRV----MYSHRDPSSRKSGTANIFIKNLDKTIDHKA 135
F + + + A++ LN + + +K + V S R + N+++KNLD +
Sbjct: 2 FESEESSNVAIEKLNGSTVGDKQLYVGKFVKKSDRILPGPDARYTNLYMKNLDLDVSEAT 181
Query: 136 LHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHF 195
L + FSSFG+I+S IA D G SKG+GFV ++ D A+ A++ +NG + K ++V
Sbjct: 182 LQEKFSSFGKIVSLVIAKDNIGMSKGFGFVNYDNPDDAKRAMEAMNGSKLGSKILYVARA 361
Query: 196 LRKQDRDNVL 205
+K +R+ +L
Sbjct: 362 QKKAEREQIL 391
Score = 55.5 bits (132), Expect = 8e-08
Identities = 25/74 (33%), Positives = 51/74 (68%)
Frame = +2
Query: 32 TSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALD 91
T+LY+ +L+++V+++ L + F+ G++VS+ + +D S G+G+VN+ NP DA RA++
Sbjct: 134 TNLYMKNLDLDVSEATLQEKFSSFGKIVSLVIAKD-NIGMSKGFGFVNYDNPDDAKRAME 310
Query: 92 VLNFTPMNNKSIRV 105
+N + + +K + V
Sbjct: 311 AMNGSKLGSKILYV 352
>BM568034 similar to GP|7673359|gb|A poly(A)-binding protein {Nicotiana
tabacum}, partial (17%)
Length = 391
Score = 117 bits (292), Expect(2) = 3e-28
Identities = 62/80 (77%), Positives = 66/80 (82%)
Frame = +2
Query: 560 VPPMPIHALATALANAPPEQQRTMLGEALYPLVDQLEHDSAAKVTGMLLEMDQPEVLHLI 619
VP + TALA APPEQQRTMLGEALYPLVDQLEHDSAAK TGMLLEMDQPEVL LI
Sbjct: 5 VPSSHEASCPTALAXAPPEQQRTMLGEALYPLVDQLEHDSAAKATGMLLEMDQPEVLPLI 184
Query: 620 ESPDALKAKVAEAMEVLRNV 639
ESP+AL +K AE M+ LRNV
Sbjct: 185 ESPNALLSKXAETMDGLRNV 244
Score = 27.3 bits (59), Expect(2) = 3e-28
Identities = 12/13 (92%), Positives = 13/13 (99%)
Frame = +1
Query: 646 SPADQLASLSLND 658
+PADQLASLSLND
Sbjct: 259 NPADQLASLSLND 297
>TC220963 similar to GB|AAA32832.1|166786|ATHPABP poly(A)-binding protein
{Arabidopsis thaliana;} , partial (9%)
Length = 522
Score = 118 bits (296), Expect = 7e-27
Identities = 77/167 (46%), Positives = 97/167 (57%), Gaps = 8/167 (4%)
Frame = +3
Query: 395 KPLYVAVAQRKEDRRARLQAQFSQMRPVAITPSVAPRMPLYPPGTPGLG-QQFMYGQGPP 453
KPLYVAVAQRKE+R+A LQAQF+Q+R + +PLY PG P L QQ YGQG P
Sbjct: 12 KPLYVAVAQRKEERKAHLQAQFAQIRAPGGMAPLPAGIPLYHPGAPRLAPQQLYYGQGTP 191
Query: 454 AMMPPQ-AGFGYQQQLVPGMRPGGGPMPSYFVP--MVQQGQQGQRPGGRRGGPGQQPPQQ 510
MPPQ AG+ +QQQ++PGMRP G P++ +P + +QGQ QR G RR G QQ
Sbjct: 192 GFMPPQPAGYSFQQQILPGMRP--GVAPNFIMPYHLQRQGQLEQRTGVRRNGNFQQ---- 353
Query: 511 VPMMQQQMLPR--QRVYRYPPGRSNIQDAPV--QNIGGGMMSYDMGG 553
+ Q QML R + +RY P N D + + G MM G
Sbjct: 354 --VQQNQMLHRNSNQGFRYMPNGRNGMDPSLVPHGLMGPMMPMPFDG 488
>TC225235 similar to GB|AAP37853.1|30725662|BT008494 At1g11650 {Arabidopsis
thaliana;} , partial (64%)
Length = 1154
Score = 107 bits (268), Expect = 1e-23
Identities = 80/293 (27%), Positives = 133/293 (45%), Gaps = 26/293 (8%)
Frame = +3
Query: 7 QHQTPAPVPAPSNG--VVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVC 64
QHQ P P+ APS + A+ +L++GDL+ ++++ LY F G+V SV+V
Sbjct: 159 QHQAPQPMWAPSAQPPLPQQPASADEVRTLWIGDLQYWMDENYLYTCFAHTGEVSSVKVI 338
Query: 65 RDLATRRSLGYGYVNFTNPQDAARALDVLN--FTPMNNKSIRVMYSHRDPSSRK----SG 118
R+ T +S GYG++ F + A R L N P +S R+ ++ R S
Sbjct: 339 RNKQTSQSEGYGFIEFNSRAGAERILQTYNGAIMPNGGQSFRLNWATFSAGERSRQDDSP 518
Query: 119 TANIFIKNLDKTIDHKALHDTF-SSFGQIMSCKIATDG-SGQSKGYGFVQFEAEDSAQNA 176
IF+ +L + L +TF + + + K+ D +G++KGYGFV+F E A
Sbjct: 519 DYTIFVGDLAADVTDYLLQETFRARYNSVKGAKVVIDRLTGRTKGYGFVRFSEESEQMRA 698
Query: 177 IDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNN----------------VYVKNLSE 220
+ ++ G+L + + + +G K K + N ++V NL
Sbjct: 699 MTEMQGVLCSTRPMRIGPASNKTPATQSQPKASYLNSQPQGSQNENDPNNTTIFVGNLDP 878
Query: 221 SFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNG 273
+ T+D L+ F YG + + K GFV F + A +A+ LNG
Sbjct: 879 NVTDDHLRQVFSQYGELVHVKI-----PAGKRCGFVQFADRSCAEEALRVLNG 1022
Score = 81.3 bits (199), Expect = 1e-15
Identities = 70/305 (22%), Positives = 142/305 (45%), Gaps = 15/305 (4%)
Frame = +3
Query: 122 IFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDG-SGQSKGYGFVQFEAEDSAQNAIDKL 180
++I +L +D L+ F+ G++ S K+ + + QS+GYGF++F + A+ +
Sbjct: 246 LWIGDLQYWMDENYLYTCFAHTGEVSSVKVIRNKQTSQSEGYGFIEFNSRAGAERILQTY 425
Query: 181 NGMLI-NDKQVF--------VGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEF 231
NG ++ N Q F G R+ D + ++V +L+ T+ L+ F
Sbjct: 426 NGAIMPNGGQSFRLNWATFSAGERSRQDDSPDY-------TIFVGDLAADVTDYLLQETF 584
Query: 232 GA-YGTITSAVLMRDA-DGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKK 289
A Y ++ A ++ D GR+K +GFV F + +A+ + G +G A K
Sbjct: 585 RARYNSVKGAKVVIDRLTGRTKGYGFVRFSEESEQMRAMTEMQGVLCSTRPMRIGPASNK 764
Query: 290 SEREQ-ELKGRFEQTVKESVVDKFQGLN--LYLKNLDDSITDEKLKEMFSEFGTITSYKV 346
+ Q + K + + + ++ N +++ NLD ++TD+ L+++FS++G + K+
Sbjct: 765 TPATQSQPKASYLNSQPQGSQNENDPNNTTIFVGNLDPNVTDDHLRQVFSQYGELVHVKI 944
Query: 347 CTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKE 406
P G + GFV F+ A AL +NG ++ + + ++ +
Sbjct: 945 ---------------PAG--KRCGFVQFADRSCAEEALRVLNGTLLGGQNVRLSWGRSPS 1073
Query: 407 DRRAR 411
+++A+
Sbjct: 1074NKQAQ 1088
Score = 48.5 bits (114), Expect = 9e-06
Identities = 29/86 (33%), Positives = 53/86 (60%)
Frame = +3
Query: 32 TSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALD 91
T+++VG+L+ NV D L +F+Q G++V V++ A +R G+V F + A AL
Sbjct: 849 TTIFVGNLDPNVTDDHLRQVFSQYGELVHVKIP---AGKRC---GFVQFADRSCAEEALR 1010
Query: 92 VLNFTPMNNKSIRVMYSHRDPSSRKS 117
VLN T + +++R+ + R PS++++
Sbjct: 1011VLNGTLLGGQNVRLSWG-RSPSNKQA 1085
>TC227320 similar to UP|Q93YF1 (Q93YF1) Nucleic acid binding protein, partial
(70%)
Length = 1724
Score = 107 bits (266), Expect = 2e-23
Identities = 81/310 (26%), Positives = 148/310 (47%), Gaps = 6/310 (1%)
Frame = +1
Query: 109 HRDPSSRKSGTAN--IFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDG-SGQSKGYGFV 165
H+ SG N I+I +L +D LH F+S G+I S K+ + +G S+GYGFV
Sbjct: 385 HQHQHQNGSGGENKTIWIGDLHHWMDENYLHRCFASTGEISSIKVIRNKQTGLSEGYGFV 564
Query: 166 QFEAEDSAQNAIDKLNGMLI-NDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTE 224
+F + +A+ + G+L+ N +Q F ++ D +++V +L+ T+
Sbjct: 565 EFYSHATAEKVLQNYAGILMPNAEQPFRLNWATFSTGDKGSDNVPDLSIFVGDLAADVTD 744
Query: 225 DDLKNEFGA-YGTITSAVLMRDAD-GRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWY 282
L F + Y ++ +A ++ DA+ GRSK +GFV F + + +A+ +NG
Sbjct: 745 SLLHETFASVYPSVKAAKVVFDANTGRSKGYGFVRFGDDNERTQAMTQMNGVYCSSRPMR 924
Query: 283 VGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTIT 342
+G A + + G T +S D +++ LD +++DE L++ FS++G I
Sbjct: 925 IGAATPRKSSGHQQGGLSNGTANQSEADS-TNTTIFVGGLDPNVSDEDLRQPFSQYGEIV 1101
Query: 343 SYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVA 402
S K+ V +G GFV F+ A AL ++NG I + + ++
Sbjct: 1102SVKI-----------------PVGKGCGFVQFANRNNAEEALQKLNGTTIGKQTVRLSWG 1230
Query: 403 QRKEDRRARL 412
+ +++ R+
Sbjct: 1231RNPANKQFRM 1260
Score = 95.1 bits (235), Expect = 9e-20
Identities = 70/265 (26%), Positives = 130/265 (48%), Gaps = 21/265 (7%)
Frame = +1
Query: 33 SLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDV 92
++++GDL ++++ L+ F G++ S++V R+ T S GYG+V F + A + L
Sbjct: 427 TIWIGDLHHWMDENYLHRCFASTGEISSIKVIRNKQTGLSEGYGFVEFYSHATAEKVLQ- 603
Query: 93 LNFT----PMNNKSIRV---MYSHRDPSSRKSGTANIFIKNLDKTIDHKALHDTFSS-FG 144
N+ P + R+ +S D S +IF+ +L + LH+TF+S +
Sbjct: 604 -NYAGILMPNAEQPFRLNWATFSTGDKGSDNVPDLSIFVGDLAADVTDSLLHETFASVYP 780
Query: 145 QIMSCKIATD-GSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQD--- 200
+ + K+ D +G+SKGYGFV+F ++ A+ ++NG+ + + + +G ++
Sbjct: 781 SVKAAKVVFDANTGRSKGYGFVRFGDDNERTQAMTQMNGVYCSSRPMRIGAATPRKSSGH 960
Query: 201 RDNVLSKTKFN---------NVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSK 251
+ LS N ++V L + +++DL+ F YG I S + K
Sbjct: 961 QQGGLSNGTANQSEADSTNTTIFVGGLDPNVSDEDLRQPFSQYGEIVSVKI-----PVGK 1125
Query: 252 CFGFVNFENAEDAAKAVEALNGKKV 276
GFV F N +A +A++ LNG +
Sbjct: 1126GCGFVQFANRNNAEEALQKLNGTTI 1200
Score = 84.0 bits (206), Expect = 2e-16
Identities = 57/175 (32%), Positives = 87/175 (49%), Gaps = 17/175 (9%)
Frame = +1
Query: 33 SLYVGDLEVNVNDSQLYDLFNQV-GQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALD 91
S++VGDL +V DS L++ F V V + +V D T RS GYG+V F + + +A+
Sbjct: 706 SIFVGDLAADVTDSLLHETFASVYPSVKAAKVVFDANTGRSKGYGFVRFGDDNERTQAMT 885
Query: 92 VLNFTPMNNKSIRV------MYSHRDPSSRKSGTAN----------IFIKNLDKTIDHKA 135
+N +++ +R+ S +GTAN IF+ LD + +
Sbjct: 886 QMNGVYCSSRPMRIGAATPRKSSGHQQGGLSNGTANQSEADSTNTTIFVGGLDPNVSDED 1065
Query: 136 LHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQV 190
L FS +G+I+S KI KG GFVQF ++A+ A+ KLNG I + V
Sbjct: 1066LRQPFSQYGEIVSVKIPV-----GKGCGFVQFANRNNAEEALQKLNGTTIGKQTV 1215
Score = 54.3 bits (129), Expect = 2e-07
Identities = 31/99 (31%), Positives = 54/99 (54%)
Frame = +1
Query: 18 SNGVVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGY 77
SNG T+++VG L+ NV+D L F+Q G++VSV++ G G+
Sbjct: 976 SNGTANQSEADSTNTTIFVGGLDPNVSDEDLRQPFSQYGEIVSVKI------PVGKGCGF 1137
Query: 78 VNFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPSSRK 116
V F N +A AL LN T + +++R+ + R+P++++
Sbjct: 1138 VQFANRNNAEEALQKLNGTTIGKQTVRLSWG-RNPANKQ 1251
>TC225237 similar to UP|Q9LEB4 (Q9LEB4) RNA Binding Protein 45, partial (79%)
Length = 1560
Score = 103 bits (257), Expect = 2e-22
Identities = 79/293 (26%), Positives = 131/293 (43%), Gaps = 26/293 (8%)
Frame = +3
Query: 7 QHQTPAPVPAPSNGVVPNV--ANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVC 64
Q Q P + APS P A+ +L++GDL+ ++++ LY F G+V SV+V
Sbjct: 225 QAQAPQAMWAPSAQPPPQQQPASADEVRTLWIGDLQYWMDENYLYTCFAHTGEVTSVKVI 404
Query: 65 RDLATRRSLGYGYVNFTNPQDAARALDVLN--FTPMNNKSIRVMYSHRDPSSRK----SG 118
R+ T +S GYG++ F + A R L N P +S R+ ++ R S
Sbjct: 405 RNKQTSQSEGYGFIEFNSRAGAERILQTYNGAIMPNGGQSFRLNWATFSAGERSRHDDSP 584
Query: 119 TANIFIKNLDKTIDHKALHDTF-SSFGQIMSCKIATDG-SGQSKGYGFVQFEAEDSAQNA 176
IF+ +L + L +TF + + + K+ D +G++KGYGFV+F E A
Sbjct: 585 DYTIFVGDLAADVTDYLLQETFRARYNSVKGAKVVIDRLTGRTKGYGFVRFSDESEQVRA 764
Query: 177 IDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNN----------------VYVKNLSE 220
+ ++ G+L + + + +G K K + N ++V NL
Sbjct: 765 MTEMQGVLCSTRPMRIGPASNKTPTTQSQPKASYQNSQPQGSQNENDPNNTTIFVGNLDP 944
Query: 221 SFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNG 273
+ T+D L+ F YG + + K GFV F + A +A+ LNG
Sbjct: 945 NVTDDHLRQVFSQYGELVHVKI-----PAGKRCGFVQFADRSCAEEALRVLNG 1088
Score = 85.9 bits (211), Expect = 5e-17
Identities = 97/412 (23%), Positives = 175/412 (41%), Gaps = 15/412 (3%)
Frame = +3
Query: 122 IFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDG-SGQSKGYGFVQFEAEDSAQNAIDKL 180
++I +L +D L+ F+ G++ S K+ + + QS+GYGF++F + A+ +
Sbjct: 312 LWIGDLQYWMDENYLYTCFAHTGEVTSVKVIRNKQTSQSEGYGFIEFNSRAGAERILQTY 491
Query: 181 NGMLI-NDKQVF--------VGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEF 231
NG ++ N Q F G R D + ++V +L+ T+ L+ F
Sbjct: 492 NGAIMPNGGQSFRLNWATFSAGERSRHDDSPDY-------TIFVGDLAADVTDYLLQETF 650
Query: 232 GA-YGTITSAVLMRDA-DGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKK 289
A Y ++ A ++ D GR+K +GFV F + + +A+ + G +G A K
Sbjct: 651 RARYNSVKGAKVVIDRLTGRTKGYGFVRFSDESEQVRAMTEMQGVLCSTRPMRIGPASNK 830
Query: 290 SEREQ-ELKGRFEQTVKESVVDKFQGLN--LYLKNLDDSITDEKLKEMFSEFGTITSYKV 346
+ Q + K ++ + + ++ N +++ NLD ++TD+ L+++FS++G + K+
Sbjct: 831 TPTTQSQPKASYQNSQPQGSQNENDPNNTTIFVGNLDPNVTDDHLRQVFSQYGELVHVKI 1010
Query: 347 CTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKE 406
P G + GFV F+ A AL +NG ++ + + ++ +
Sbjct: 1011---------------PAG--KRCGFVQFADRSCAEEALRVLNGTLLGGQNVRLSWGRSPS 1139
Query: 407 DRRARLQAQFSQMRPVAITPSVAPRMPLYPPGTPGLGQQFMYGQGPPAMMPPQAGFGYQQ 466
+++A QA +Q G G G + YG A GY+
Sbjct: 1140NKQA--QADPNQWN-----------------GGAGSGGGY-YG---------YAAQGYEN 1232
Query: 467 QLVPGMRPGGGPMPSYFVPMVQQGQQGQRPGGRRGGPGQQPPQQVPMMQQQM 518
G P G P+ + G G PG QPPQQ QQQ+
Sbjct: 1233Y---GYAPAAGQDPNMY-------------GSYPGYPGYQPPQQ----QQQI 1328
Score = 48.5 bits (114), Expect = 9e-06
Identities = 29/86 (33%), Positives = 53/86 (60%)
Frame = +3
Query: 32 TSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALD 91
T+++VG+L+ NV D L +F+Q G++V V++ A +R G+V F + A AL
Sbjct: 915 TTIFVGNLDPNVTDDHLRQVFSQYGELVHVKIP---AGKRC---GFVQFADRSCAEEALR 1076
Query: 92 VLNFTPMNNKSIRVMYSHRDPSSRKS 117
VLN T + +++R+ + R PS++++
Sbjct: 1077VLNGTLLGGQNVRLSWG-RSPSNKQA 1151
>TC223000 similar to GB|AAG02117.1|9930616|AF293840 poly(A) binding protein
{Arabidopsis thaliana;} , partial (11%)
Length = 775
Score = 103 bits (256), Expect = 3e-22
Identities = 73/152 (48%), Positives = 90/152 (59%), Gaps = 10/152 (6%)
Frame = +2
Query: 517 QMLPRQ--RVYRYPPGRSNIQDAPVQNIG--GGMMSYDMGG-----LPLRDVVPPMPIHA 567
QML R + +RY P N D + G G MM G P + P
Sbjct: 44 QMLHRNSNQGFRYMPNGRNGMDPSLVPHGLMGPMMPMPFDGSGVTAAPNDNQRPGALSTT 223
Query: 568 LATALANAPPEQQRTMLGEALYPLVDQLEHDS-AAKVTGMLLEMDQPEVLHLIESPDALK 626
LA+ALA+A PE QR MLGE LYPLV++L + AKVTGMLLEMDQ EV++LIESP+ LK
Sbjct: 224 LASALASATPENQRMMLGEHLYPLVERLAPNQYTAKVTGMLLEMDQSEVINLIESPEDLK 403
Query: 627 AKVAEAMEVLRNVSQQQGNSPADQLASLSLND 658
KV+EAM+VL V+ S DQL S+SLN+
Sbjct: 404 TKVSEAMQVLHEVA---SGSEVDQLGSMSLNE 490
>TC214546 similar to PIR|T01932|T01932 RNA binding protein homolog - common
tobacco (fragment) {Nicotiana tabacum;} , partial (68%)
Length = 1756
Score = 95.1 bits (235), Expect = 9e-20
Identities = 79/311 (25%), Positives = 142/311 (45%), Gaps = 45/311 (14%)
Frame = +3
Query: 8 HQTPAPVPAPSNG-----------VVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVG 56
H P P P P A+ ++++GDL ++++ L++ F G
Sbjct: 384 HYPPPPPPPPPQSSHHHHHKQAAAAAAAAASSDEIRTVWLGDLHHWMDENYLHNCFAHTG 563
Query: 57 QVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDVLNFTPMNN--KSIRVMYSHRDPSS 114
+VVS +V R+ T +S GYG+V F + A + L N T M N ++ R+ ++
Sbjct: 564 EVVSAKVIRNKQTGQSEGYGFVEFYSRATAEKVLQNYNGTMMPNTDQAFRLNWATFSAGE 743
Query: 115 RKSGTA----NIFIKNLDKTIDHKALHDTFSS-FGQIMSCKIATD-GSGQSKGYGFVQFE 168
R+S A +IF+ +L + L +TF+ + I K+ D +G+SKGYGFV+F
Sbjct: 744 RRSSDATSDLSIFVGDLAIDVTDAMLQETFAGRYSSIKGAKVVIDSNTGRSKGYGFVRFG 923
Query: 169 AEDSAQNAIDKLNGMLINDKQVFVG--------HFLRKQDRDNVL--------------- 205
E+ A+ ++NG+ + + + +G F ++ V+
Sbjct: 924 DENERTRAMTEMNGVYCSSRPMRIGVATPKKTYGFQQQYSSQAVVLAGGHSANGAVAQGS 1103
Query: 206 -SKTKFNN--VYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAE 262
S+ NN ++V L +++DL+ F +G + S + K GFV F + +
Sbjct: 1104HSEGDINNTTIFVGGLDSDTSDEDLRQPFLQFGEVVSVKI-----PVGKGCGFVQFADRK 1268
Query: 263 DAAKAVEALNG 273
+A +A++ LNG
Sbjct: 1269NAEEAIQGLNG 1301
Score = 91.7 bits (226), Expect = 1e-18
Identities = 75/289 (25%), Positives = 136/289 (46%), Gaps = 18/289 (6%)
Frame = +3
Query: 122 IFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDG-SGQSKGYGFVQFEAEDSAQNAIDKL 180
+++ +L +D LH+ F+ G+++S K+ + +GQS+GYGFV+F + +A+ +
Sbjct: 495 VWLGDLHHWMDENYLHNCFAHTGEVVSAKVIRNKQTGQSEGYGFVEFYSRATAEKVLQNY 674
Query: 181 NG-MLINDKQVFVGHFLRKQDRDNVLSK-TKFNNVYVKNLSESFTEDDLKNEF-GAYGTI 237
NG M+ N Q F ++ + S T +++V +L+ T+ L+ F G Y +I
Sbjct: 675 NGTMMPNTDQAFRLNWATFSAGERRSSDATSDLSIFVGDLAIDVTDAMLQETFAGRYSSI 854
Query: 238 TSAVLMRDAD-GRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKA---------Q 287
A ++ D++ GRSK +GFV F + + +A+ +NG +G A Q
Sbjct: 855 KGAKVVIDSNTGRSKGYGFVRFGDENERTRAMTEMNGVYCSSRPMRIGVATPKKTYGFQQ 1034
Query: 288 KKSEREQELKGRFEQTVKESVVDKFQG----LNLYLKNLDDSITDEKLKEMFSEFGTITS 343
+ S + L G + +G +++ LD +DE L++ F +FG + S
Sbjct: 1035QYSSQAVVLAGGHSANGAVAQGSHSEGDINNTTIFVGGLDSDTSDEDLRQPFLQFGEVVS 1214
Query: 344 YKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMI 392
K+ V +G GFV F+ + A A+ +NG +I
Sbjct: 1215VKI-----------------PVGKGCGFVQFADRKNAEEAIQGLNGTVI 1310
Score = 71.2 bits (173), Expect = 1e-12
Identities = 55/189 (29%), Positives = 80/189 (42%), Gaps = 31/189 (16%)
Frame = +3
Query: 33 SLYVGDLEVNVNDSQLYDLF-NQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALD 91
S++VGDL ++V D+ L + F + + +V D T RS GYG+V F + + RA+
Sbjct: 774 SIFVGDLAIDVTDAMLQETFAGRYSSIKGAKVVIDSNTGRSKGYGFVRFGDENERTRAMT 953
Query: 92 VLN-------------FTPMNNKSIRVMYSHRD--------------PSSRKSGTAN--- 121
+N TP + YS + S G N
Sbjct: 954 EMNGVYCSSRPMRIGVATPKKTYGFQQQYSSQAVVLAGGHSANGAVAQGSHSEGDINNTT 1133
Query: 122 IFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLN 181
IF+ LD + L F FG+++S KI KG GFVQF +A+ AI LN
Sbjct: 1134IFVGGLDSDTSDEDLRQPFLQFGEVVSVKIPV-----GKGCGFVQFADRKNAEEAIQGLN 1298
Query: 182 GMLINDKQV 190
G +I + V
Sbjct: 1299GTVIGKQTV 1325
Score = 65.1 bits (157), Expect = 1e-10
Identities = 53/202 (26%), Positives = 93/202 (45%), Gaps = 1/202 (0%)
Frame = +3
Query: 206 SKTKFNNVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDAD-GRSKCFGFVNFENAEDA 264
S + V++ +L E+ L N F G + SA ++R+ G+S+ +GFV F + A
Sbjct: 474 SSDEIRTVWLGDLHHWMDENYLHNCFAHTGEVVSAKVIRNKQTGQSEGYGFVEFYSRATA 653
Query: 265 AKAVEALNGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDD 324
K ++ NG + + +++ L + D L++++ +L
Sbjct: 654 EKVLQNYNGTMMPN-----------TDQAFRLNWATFSAGERRSSDATSDLSIFVGDLAI 800
Query: 325 SITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRAL 384
+TD L+E F+ G +S K +V++ G S+G GFV F E +RA+
Sbjct: 801 DVTDAMLQETFA--GRYSSIK--------GAKVVIDSNTGRSKGYGFVRFGDENERTRAM 950
Query: 385 GEMNGKMIVSKPLYVAVAQRKE 406
EMNG S+P+ + VA K+
Sbjct: 951 TEMNGVYCSSRPMRIGVATPKK 1016
Score = 42.7 bits (99), Expect = 5e-04
Identities = 25/84 (29%), Positives = 47/84 (55%)
Frame = +3
Query: 32 TSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALD 91
T+++VG L+ + +D L F Q G+VVSV++ G G+V F + ++A A+
Sbjct: 1128 TTIFVGGLDSDTSDEDLRQPFLQFGEVVSVKI------PVGKGCGFVQFADRKNAEEAIQ 1289
Query: 92 VLNFTPMNNKSIRVMYSHRDPSSR 115
LN T + +++R+ + R P ++
Sbjct: 1290 GLNGTVIGKQTVRLSWG-RSPGNK 1358
>BM308944
Length = 429
Score = 92.0 bits (227), Expect = 7e-19
Identities = 50/114 (43%), Positives = 72/114 (62%)
Frame = +3
Query: 35 YVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDVLN 94
YVG+L+ + + L++LF Q G VV+V V +D T + GYG+V F + +DA A+ VLN
Sbjct: 90 YVGNLDPQICEELLWELFVQAGPVVNVYVPKDRVTNQHQGYGFVEFRSEEDADYAIKVLN 269
Query: 95 FTPMNNKSIRVMYSHRDPSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMS 148
+ K IRV + +D S G AN+FI NLD +D K L+DTFS+FG I++
Sbjct: 270 MIKLYGKPIRVNKASQDKKSLDVG-ANLFIGNLDPDVDEKLLYDTFSAFGVIVT 428
Score = 51.2 bits (121), Expect = 1e-06
Identities = 36/128 (28%), Positives = 63/128 (49%), Gaps = 1/128 (0%)
Frame = +3
Query: 113 SSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDG-SGQSKGYGFVQFEAED 171
++ ++ A ++ NLD I + L + F G +++ + D + Q +GYGFV+F +E+
Sbjct: 60 AAERNQDATAYVGNLDPQICEELLWELFVQAGPVVNVYVPKDRVTNQHQGYGFVEFRSEE 239
Query: 172 SAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEF 231
A AI LN + + K + V QD+ S N+++ NL E L + F
Sbjct: 240 DADYAIKVLNMIKLYGKPIRVNK--ASQDKK---SLDVGANLFIGNLDPDVDEKLLYDTF 404
Query: 232 GAYGTITS 239
A+G I +
Sbjct: 405 SAFGVIVT 428
Score = 46.2 bits (108), Expect = 5e-05
Identities = 37/131 (28%), Positives = 60/131 (45%), Gaps = 1/131 (0%)
Frame = +3
Query: 214 YVKNLSESFTEDDLKNEFGAYGTITSAVLMRD-ADGRSKCFGFVNFENAEDAAKAVEALN 272
YV NL E+ L F G + + + +D + + +GFV F + EDA A++ LN
Sbjct: 90 YVGNLDPQICEELLWELFVQAGPVVNVYVPKDRVTNQHQGYGFVEFRSEEDADYAIKVLN 269
Query: 273 GKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKLK 332
K+ + V KA S+ ++ L G NL++ NLD + ++ L
Sbjct: 270 MIKLYGKPIRVNKA---SQDKKSLD---------------VGANLFIGNLDPDVDEKLLY 395
Query: 333 EMFSEFGTITS 343
+ FS FG I +
Sbjct: 396 DTFSAFGVIVT 428
Score = 35.0 bits (79), Expect = 0.11
Identities = 25/99 (25%), Positives = 43/99 (43%)
Frame = +3
Query: 306 ESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGV 365
+ ++ Q Y+ NLD I +E L E+F + G + ++ V
Sbjct: 54 QHAAERNQDATAYVGNLDPQICEELLWELFVQAGPV-----------VNVYVPKDRVTNQ 200
Query: 366 SRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQR 404
+G GFV F + E+A A+ +N + KP+ V A +
Sbjct: 201 HQGYGFVEFRSEEDADYAIKVLNMIKLYGKPIRVNKASQ 317
>AW756264 weakly similar to GP|13560783|gb| poly(A)-binding protein {Daucus
carota}, partial (12%)
Length = 355
Score = 89.7 bits (221), Expect = 4e-18
Identities = 57/110 (51%), Positives = 65/110 (58%), Gaps = 4/110 (3%)
Frame = +2
Query: 538 PVQNIGGGMMSYDM---GGLPLRDVV-PPMPIHALATALANAPPEQQRTMLGEALYPLVD 593
P + GMMS + P+R P P A ATA A APPEQQRTMLGEA +P D
Sbjct: 2 PPSGVSCGMMSRPLRRWAARPIRHAEGQPRPKQAEATAQAKAPPEQQRTMLGEARHPRGD 181
Query: 594 QLEHDSAAKVTGMLLEMDQPEVLHLIESPDALKAKVAEAMEVLRNVSQQQ 643
Q EHD+AAK TG E DQPE E+PDA KAK AE + RN +QQQ
Sbjct: 182 QREHDAAAKGTGTQQEKDQPERGQRREAPDARKAKGAETTDGERNGAQQQ 331
>TC214650 similar to UP|Q9LJH8 (Q9LJH8) RNA binding protein nucleolysin;
oligouridylate binding protein (AT3g14100/MAG2_5),
partial (93%)
Length = 1684
Score = 89.4 bits (220), Expect = 5e-18
Identities = 81/310 (26%), Positives = 131/310 (42%), Gaps = 17/310 (5%)
Frame = +2
Query: 105 VMYSHRDPSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGF 164
++ + P S ++++ N+ + L + FS+ G + CK+ + YGF
Sbjct: 299 ILSGNLPPGFDSSSCRSVYVGNIHPQVTDSLLQELFSTAGALEGCKLIRK---EKSSYGF 469
Query: 165 VQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTE 224
V + SA AI LNG I + + V R++ N++V +LS T+
Sbjct: 470 VDYFDRSSAAFAIVTLNGRNIFGQPIKVNWAYASSQREDTSGHF---NIFVGDLSPEVTD 640
Query: 225 DDLKNEFGAYGTITSAVLMRDAD-GRSKCFGFVNFENAEDAAKAVEALNGKKVDDEE--- 280
L F Y + + A +M D GRS+ FGFV+F N +DA A+ L GK + +
Sbjct: 641 ATLYACFSVYPSCSDARVMWDQKTGRSRGFGFVSFRNQQDAQSAINDLTGKWLGSRQIRC 820
Query: 281 -WYVGKAQKKSEREQ-------ELKGRFEQTVKESVVDKF-----QGLNLYLKNLDDSIT 327
W A E++ EL + +E+ D Q +Y+ NL +T
Sbjct: 821 NWATKGASASDEKQTSDSRSVVELTNGSSEDGQETTNDDTPEKNPQYTTVYVGNLAPEVT 1000
Query: 328 DEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEM 387
L + F T D+RV RD +G GFV +ST EA+ A+
Sbjct: 1001SVDLHQHFHSLNAGT---------IEDVRV-QRD-----KGFGFVRYSTHAEAALAIQMG 1135
Query: 388 NGKMIVSKPL 397
N +++ KP+
Sbjct: 1136NARILFGKPI 1165
Score = 57.8 bits (138), Expect = 2e-08
Identities = 47/193 (24%), Positives = 76/193 (39%), Gaps = 35/193 (18%)
Frame = +2
Query: 33 SLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDV 92
+++VGDL V D+ LY F+ RV D T RS G+G+V+F N QDA A++
Sbjct: 602 NIFVGDLSPEVTDATLYACFSVYPSCSDARVMWDQKTGRSRGFGFVSFRNQQDAQSAIND 781
Query: 93 LNFTPMNNKSIRVMYSHR---------------------------------DPSSRKSGT 119
L + ++ IR ++ + D +
Sbjct: 782 LTGKWLGSRQIRCNWATKGASASDEKQTSDSRSVVELTNGSSEDGQETTNDDTPEKNPQY 961
Query: 120 ANIFIKNLDKTIDHKALHDTFSSF--GQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAI 177
+++ NL + LH F S G I ++ D KG+GFV++ A AI
Sbjct: 962 TTVYVGNLAPEVTSVDLHQHFHSLNAGTIEDVRVQRD-----KGFGFVRYSTHAEAALAI 1126
Query: 178 DKLNGMLINDKQV 190
N ++ K +
Sbjct: 1127QMGNARILFGKPI 1165
Score = 33.5 bits (75), Expect = 0.31
Identities = 25/124 (20%), Positives = 52/124 (41%), Gaps = 3/124 (2%)
Frame = +2
Query: 316 NLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFS 375
++Y+ N+ +TD L+E+FS G + K+ I ++ + GFV +
Sbjct: 347 SVYVGNIHPQVTDSLLQELFSTAGALEGCKL-----------IRKEKSSY----GFVDYF 481
Query: 376 TPEEASRALGEMNGKMIVSKPL---YVAVAQRKEDRRARLQAQFSQMRPVAITPSVAPRM 432
A+ A+ +NG+ I +P+ + + ++ED + P ++
Sbjct: 482 DRSSAAFAIVTLNGRNIFGQPIKVNWAYASSQREDTSGHFNIFVGDLSPEVTDATLYACF 661
Query: 433 PLYP 436
+YP
Sbjct: 662 SVYP 673
>TC210229 similar to UP|O57336 (O57336) Polyadenylate binding protein,
partial (11%)
Length = 1101
Score = 86.7 bits (213), Expect = 3e-17
Identities = 83/234 (35%), Positives = 108/234 (45%), Gaps = 25/234 (10%)
Frame = +3
Query: 430 PRMPLYPPGTPGLGQQFMYGQGPPAMMPPQAGFGYQQQLVPGMRPGGG------PMPSYF 483
P + P G P + G + +PP+AG YQ +P MRPG G P S+
Sbjct: 24 PSTAIIPGGYP---PYYYAATGVISHVPPRAGLMYQH--LP-MRPGWGANGFALPARSFQ 185
Query: 484 ---VPMVQQG--QQGQRPGGRRGGP--------------GQQPPQQVPMMQQQMLPRQRV 524
VP V Q Q G G P QQ Q V Q Q
Sbjct: 186 QSPVPAVSNNTRQHRQNRGRLNGNPIAQGNTHSVTYLPQAQQISQPVISSSTQQRTGQAK 365
Query: 525 YRYPPGRSNIQDAPVQNIGGGMMSYDMGGLPLRDVVPPMPIHALATALANAPPEQQRTML 584
Y P GR +++ G S G R +H++ LA A PEQQ+ +L
Sbjct: 366 Y-LPSGRQ-------RDMEKGSGSSSSGSNSGRGSQGSEMLHSM---LAGAAPEQQKEIL 512
Query: 585 GEALYPLVDQLEHDSAAKVTGMLLEMDQPEVLHLIESPDALKAKVAEAMEVLRN 638
GE LY LV +L+ AAK+TGMLLEMD E+L L+ESP++L AKV EA++VL+N
Sbjct: 513 GEHLYMLVHKLKPTLAAKITGMLLEMDNGELLLLLESPESLSAKVEEAVQVLKN 674
>TC205913 weakly similar to UP|Q9LEB4 (Q9LEB4) RNA Binding Protein 45,
partial (55%)
Length = 886
Score = 86.7 bits (213), Expect = 3e-17
Identities = 72/275 (26%), Positives = 128/275 (46%), Gaps = 28/275 (10%)
Frame = +1
Query: 33 SLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDV 92
+L++GDL+ V++S L F G+VVS+++ R+ T + GYG+V F + A L
Sbjct: 67 TLWIGDLQYWVDESYLSQCFAHNGEVVSIKIIRNKLTGQPEGYGFVEFVSHASAEAFLRT 246
Query: 93 LNFT--PMNNKSIRVMYSHRDPSSRKSGTANIFIKNLDKTIDHKALHDTF-SSFGQIMSC 149
N P ++ R+ ++ S +IF+ +L + L +TF + + +
Sbjct: 247 YNGAQMPGTEQTFRLNWASFGDSGPDH---SIFVGDLAPDVTDFLLQETFRAHYPSVKGA 417
Query: 150 KIATD-GSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDR------- 201
K+ TD +G+SKGYGFV+F E A+ ++NG+ + + + + K++
Sbjct: 418 KVVTDPATGRSKGYGFVKFADEAQRNRAMTEMNGVYCSTRPMRISAATPKKNASFQHQYA 597
Query: 202 ---------------DNVLSKTKFNN--VYVKNLSESFTEDDLKNEFGAYGTITSAVLMR 244
V + NN V + NL + TE++LK F +G I VL++
Sbjct: 598 PPKAMYQFPAYSAPVSAVAPENDVNNTTVCIGNLDLNVTEEELKQTFMQFGDI---VLVK 768
Query: 245 DADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDE 279
G K +G+V F A A++ + GK + +
Sbjct: 769 IYAG--KGYGYVQFGTRVSAEDAIQRMQGKVIGQQ 867
Score = 77.4 bits (189), Expect = 2e-14
Identities = 73/294 (24%), Positives = 131/294 (43%), Gaps = 15/294 (5%)
Frame = +1
Query: 122 IFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDG-SGQSKGYGFVQFEAEDSAQNAIDKL 180
++I +L +D L F+ G+++S KI + +GQ +GYGFV+F + SA+ +
Sbjct: 70 LWIGDLQYWVDESYLSQCFAHNGEVVSIKIIRNKLTGQPEGYGFVEFVSHASAEAFLRTY 249
Query: 181 NG-MLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGA-YGTIT 238
NG + +Q F ++ D ++++V +L+ T+ L+ F A Y ++
Sbjct: 250 NGAQMPGTEQTFRLNWASFGDSG------PDHSIFVGDLAPDVTDFLLQETFRAHYPSVK 411
Query: 239 SAVLMRD-ADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSEREQEL- 296
A ++ D A GRSK +GFV F + +A+ +NG + A K +
Sbjct: 412 GAKVVTDPATGRSKGYGFVKFADEAQRNRAMTEMNGVYCSTRPMRISAATPKKNASFQHQ 591
Query: 297 ----KGRFEQTVKESVVDKFQGLN------LYLKNLDDSITDEKLKEMFSEFGTITSYKV 346
K ++ + V N + + NLD ++T+E+LK+ F +FG I K+
Sbjct: 592 YAPPKAMYQFPAYSAPVSAVAPENDVNNTTVCIGNLDLNVTEEELKQTFMQFGDIVLVKI 771
Query: 347 CTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVA 400
+G G+V F T A A+ M GK+I + + ++
Sbjct: 772 Y-----------------AGKGYGYVQFGTRVSAEDAIQRMQGKVIGQQVIQIS 882
Score = 72.4 bits (176), Expect = 6e-13
Identities = 58/187 (31%), Positives = 81/187 (43%), Gaps = 29/187 (15%)
Frame = +1
Query: 33 SLYVGDLEVNVNDSQLYDLFN-QVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALD 91
S++VGDL +V D L + F V +V D AT RS GYG+V F + RA+
Sbjct: 328 SIFVGDLAPDVTDFLLQETFRAHYPSVKGAKVVTDPATGRSKGYGFVKFADEAQRNRAMT 507
Query: 92 VLN-------------FTPMNNKSI-------RVMYSHRDPSSRKSGTA--------NIF 123
+N TP N S + MY S+ S A +
Sbjct: 508 EMNGVYCSTRPMRISAATPKKNASFQHQYAPPKAMYQFPAYSAPVSAVAPENDVNNTTVC 687
Query: 124 IKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGM 183
I NLD + + L TF FG I+ KI KGYG+VQF SA++AI ++ G
Sbjct: 688 IGNLDLNVTEEELKQTFMQFGDIVLVKIYA-----GKGYGYVQFGTRVSAEDAIQRMQGK 852
Query: 184 LINDKQV 190
+I + +
Sbjct: 853 VIGQQVI 873
Score = 46.6 bits (109), Expect = 4e-05
Identities = 52/231 (22%), Positives = 97/231 (41%), Gaps = 6/231 (2%)
Frame = +1
Query: 213 VYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDA-DGRSKCFGFVNFENAEDAAKAVEAL 271
+++ +L E L F G + S ++R+ G+ + +GFV F + A +
Sbjct: 70 LWIGDLQYWVDESYLSQCFAHNGEVVSIKIIRNKLTGQPEGYGFVEFVSHASAEAFLRTY 249
Query: 272 NGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVK---ESVVDKFQGLNLYLKNLDDSITD 328
NG ++ E QT + S D ++++ +L +TD
Sbjct: 250 NGAQMPGTE---------------------QTFRLNWASFGDSGPDHSIFVGDLAPDVTD 366
Query: 329 EKLKEMF-SEFGTITSYKVCTMILFTDIRVIMRDP-NGVSRGSGFVAFSTPEEASRALGE 386
L+E F + + ++ KV T DP G S+G GFV F+ + +RA+ E
Sbjct: 367 FLLQETFRAHYPSVKGAKVVT------------DPATGRSKGYGFVKFADEAQRNRAMTE 510
Query: 387 MNGKMIVSKPLYVAVAQRKEDRRARLQAQFSQMRPVAITPSVAPRMPLYPP 437
MNG ++P+ ++ A K + A Q Q++ + + P+ + + P
Sbjct: 511 MNGVYCSTRPMRISAATPK--KNASFQHQYAPPKAMYQFPAYSAPVSAVAP 657
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.134 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,813,776
Number of Sequences: 63676
Number of extensions: 311902
Number of successful extensions: 2960
Number of sequences better than 10.0: 306
Number of HSP's better than 10.0 without gapping: 2268
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2738
length of query: 659
length of database: 12,639,632
effective HSP length: 103
effective length of query: 556
effective length of database: 6,081,004
effective search space: 3381038224
effective search space used: 3381038224
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148486.10


