

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148485.4 - phase: 0
(449 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
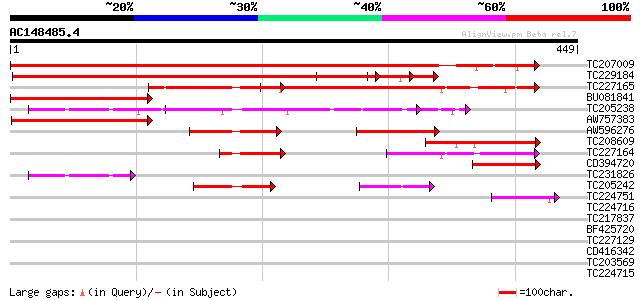
Score E
Sequences producing significant alignments: (bits) Value
TC207009 similar to UP|Q9FGL9 (Q9FGL9) Polypyrimidine tract-bind... 517 e-147
TC229184 similar to UP|O82472 (O82472) Polypyrimidine tract-bind... 493 e-140
TC227165 similar to UP|Q9FGL9 (Q9FGL9) Polypyrimidine tract-bind... 195 4e-50
BU081841 homologue to GP|9757999|dbj polypyrimidine tract-bindin... 174 6e-44
TC205238 similar to UP|Q6ICX4 (Q6ICX4) At1g43190, partial (98%) 155 5e-38
AW757383 similar to GP|9757999|dbj polypyrimidine tract-binding ... 141 7e-34
AW596276 118 6e-27
TC208609 similar to UP|O82472 (O82472) Polypyrimidine tract-bind... 111 6e-25
TC227164 similar to UP|Q9FGL9 (Q9FGL9) Polypyrimidine tract-bind... 80 2e-15
CD394720 68 7e-12
TC231826 similar to UP|Q6ICX4 (Q6ICX4) At1g43190, partial (20%) 56 3e-08
TC205242 similar to UP|Q6ICX4 (Q6ICX4) At1g43190, partial (69%) 52 7e-07
TC224751 similar to UP|Q6V8K5 (Q6V8K5) GPRP, partial (72%) 44 1e-04
TC224716 similar to GB|AAP21369.1|30102902|BT006561 At4g19200 {A... 40 0.002
TC217837 similar to PIR|S65780|S65780 glycine/proline-rich prote... 39 0.004
BF425720 39 0.004
TC227129 similar to UP|Q6V8K5 (Q6V8K5) GPRP, partial (62%) 37 0.018
CD416342 similar to SP|P42698|D111_ DNA-damage-repair/toleration... 37 0.018
TC203569 similar to GB|AAO63422.1|28950997|BT005358 At1g03457 {A... 37 0.024
TC224715 homologue to UP|Q39882 (Q39882) Nodulin-26, partial (98%) 35 0.069
>TC207009 similar to UP|Q9FGL9 (Q9FGL9) Polypyrimidine tract-binding RNA
transport protein-like, partial (80%)
Length = 1621
Score = 517 bits (1332), Expect = e-147
Identities = 264/427 (61%), Positives = 315/427 (72%), Gaps = 8/427 (1%)
Frame = +2
Query: 1 MSSSKQQQQFRCTQIPSKVLHFRNLPWECSDEELADLCSPFGKVMSTKCNVGVNQNQAFV 60
M+S Q QFR TQ PSKVLH RNLPWEC++EEL +L PFGKV++TKCNVG N+NQAF+
Sbjct: 146 MASVSSQPQFRYTQPPSKVLHLRNLPWECTEEELIELGKPFGKVVNTKCNVGANRNQAFI 325
Query: 61 QFAELNQAISMVSYYASSSEPAQVRGKTVYIQYSNRHHIVNSNSPGDIPGNVLLVTIEGV 120
+FA+LNQAI+M+SYYASSSEPAQVRGKTVY+QYSNR IVN+ + D+PGNVLLVTIEG
Sbjct: 326 EFADLNQAIAMISYYASSSEPAQVRGKTVYLQYSNRQEIVNNKTTADVPGNVLLVTIEGA 505
Query: 121 EAGDVSIDVIHLVFSAFGFVHKLATFEKTAGFQALIQYTDAETAASAKDSLDGRSIPRYL 180
+A VSIDV+HLVFSAFGFVHK+ TFEKTAGFQAL+Q++DAETA SAKD+LDGRSIPRYL
Sbjct: 506 DARLVSIDVLHLVFSAFGFVHKITTFEKTAGFQALVQFSDAETATSAKDALDGRSIPRYL 685
Query: 181 LPEHVGACNLRISYSAHRDLNIKFQSNRSRDYTNPMLPVNQAAIDSALQPAIGPDGKRKE 240
LPEH+G C LRI+YS H DL++KFQS+RSRDYTNP LPV +A++ + Q +G DGKR E
Sbjct: 686 LPEHMGPCTLRITYSGHSDLSVKFQSHRSRDYTNPYLPVAPSAVEGSGQAMVGLDGKRLE 865
Query: 241 HKSNVLLATIENMQYAVPLDVLHSVFSAFGFVQKVAMFDKNGHTHALIQYPDLTIAAAAK 300
+SNVLLA+IENMQYAV LDVLH VFSAFG VQK+AMFDKNG ALIQ+PD A AK
Sbjct: 866 AESNVLLASIENMQYAVTLDVLHMVFSAFGPVQKIAMFDKNGGLQALIQFPDTQTAVVAK 1045
Query: 301 ETLEGHCIYDGGYCKLHLTYSRHTDLNVKAFSDKSRDYTVLDPSLHAAQAPAWQTTQAAT 360
E LEGHCIYDGG+CKLH++YSRHTDL++K +D+SRDYT+ T A
Sbjct: 1046EALEGHCIYDGGFCKLHISYSRHTDLSIKVNNDRSRDYTI-------------PNTPAVN 1186
Query: 361 MYSGSMGQ--MPSWDPNQQEVTQSYLSAPGTFPSGQAAPPFP------GYSPAAVPPAGA 412
+ +GQ +P P QQ S + GT P P Y P+ P
Sbjct: 1187VQPSILGQQSVPMMGPPQQPYNGSQ-AGWGTAPPATTVQSMPMQMHNNVYMPSGTMPQQM 1363
Query: 413 SPHSHMP 419
+P P
Sbjct: 1364APGMQFP 1384
>TC229184 similar to UP|O82472 (O82472) Polypyrimidine tract-binding protein
homolog, partial (76%)
Length = 1109
Score = 493 bits (1269), Expect = e-140
Identities = 239/291 (82%), Positives = 266/291 (91%)
Frame = +3
Query: 3 SSKQQQQFRCTQIPSKVLHFRNLPWECSDEELADLCSPFGKVMSTKCNVGVNQNQAFVQF 62
S+ Q QFR TQ PSKVLH RNLPWECS+EEL +LC PFGK+++TKCNVG N+NQAFV+F
Sbjct: 135 STSSQHQFRYTQTPSKVLHLRNLPWECSEEELRELCRPFGKIVNTKCNVGANRNQAFVEF 314
Query: 63 AELNQAISMVSYYASSSEPAQVRGKTVYIQYSNRHHIVNSNSPGDIPGNVLLVTIEGVEA 122
+LNQAISMVSYYASSSEPA VRGKTVYIQYSNRH IVN+ SPGDIPGNVLLVTIEGVEA
Sbjct: 315 VDLNQAISMVSYYASSSEPAMVRGKTVYIQYSNRHEIVNNKSPGDIPGNVLLVTIEGVEA 494
Query: 123 GDVSIDVIHLVFSAFGFVHKLATFEKTAGFQALIQYTDAETAASAKDSLDGRSIPRYLLP 182
GDVSIDVIHLVFSAFGFVHK+ATFEKTAGFQALIQ+TDAETA+SA+D+LDGRSIPRYLLP
Sbjct: 495 GDVSIDVIHLVFSAFGFVHKIATFEKTAGFQALIQFTDAETASSARDALDGRSIPRYLLP 674
Query: 183 EHVGACNLRISYSAHRDLNIKFQSNRSRDYTNPMLPVNQAAIDSALQPAIGPDGKRKEHK 242
HVG+CNLRISYSAH+DLNIKFQSNRSRDYTNPMLPVN AI+ A+Q A+GPDGKRKE
Sbjct: 675 AHVGSCNLRISYSAHKDLNIKFQSNRSRDYTNPMLPVNYTAIEGAVQTAVGPDGKRKEPD 854
Query: 243 SNVLLATIENMQYAVPLDVLHSVFSAFGFVQKVAMFDKNGHTHALIQYPDL 293
SNVLLA+IENMQYAV +DVLH+VFSAFG VQK+A+F+KNG T ALIQYP +
Sbjct: 855 SNVLLASIENMQYAVTVDVLHTVFSAFGTVQKIAIFEKNGQTQALIQYPGI 1007
Score = 90.1 bits (222), Expect = 2e-18
Identities = 49/103 (47%), Positives = 66/103 (63%), Gaps = 7/103 (6%)
Frame = +3
Query: 244 NVLLATIENMQYA-VPLDVLHSVFSAFGFVQKVAMFDKNGHTHALIQYPDLTIAAAAKET 302
NVLL TIE ++ V +DV+H VFSAFGFV K+A F+K ALIQ+ D A++A++
Sbjct: 459 NVLLVTIEGVEAGDVSIDVIHLVFSAFGFVHKIATFEKTAGFQALIQFTDAETASSARDA 638
Query: 303 LEGHCI------YDGGYCKLHLTYSRHTDLNVKAFSDKSRDYT 339
L+G I G C L ++YS H DLN+K S++SRDYT
Sbjct: 639 LDGRSIPRYLLPAHVGSCNLRISYSAHKDLNIKFQSNRSRDYT 767
Score = 55.1 bits (131), Expect = 6e-08
Identities = 24/37 (64%), Positives = 29/37 (77%)
Frame = +1
Query: 284 THALIQYPDLTIAAAAKETLEGHCIYDGGYCKLHLTY 320
T LI + + A+AA+E LEGHCIYDGGYCKLHL+Y
Sbjct: 997 TLVLIIFNYIITASAAREALEGHCIYDGGYCKLHLSY 1107
>TC227165 similar to UP|Q9FGL9 (Q9FGL9) Polypyrimidine tract-binding RNA
transport protein-like, partial (31%)
Length = 1123
Score = 195 bits (495), Expect = 4e-50
Identities = 115/236 (48%), Positives = 148/236 (61%), Gaps = 15/236 (6%)
Frame = +2
Query: 199 DLNIKFQSNRSRDYT---NPMLPVNQAAIDSALQPAIGPDGKRKEHKSNVLLATIENMQY 255
D+ IK Q ++S + NP LPV Q+A++ + QP +G DGKR E +SNVLLA+IENMQY
Sbjct: 50 DIQIKCQVSKSTEAETTINPYLPVAQSAMEGSGQPMVGLDGKRLEAESNVLLASIENMQY 229
Query: 256 AVPLDVLHSVFSAFGFVQKVAMFDKNGHTHALIQYPDLTIAAAAKETLEGHCIYDGGYCK 315
V LDVLH VFSAFG VQK+AMFDKNG ALIQYPD+ A AKETLEGHCIYDGG+CK
Sbjct: 230 TVTLDVLHMVFSAFGPVQKIAMFDKNGGLQALIQYPDIQTAVVAKETLEGHCIYDGGFCK 409
Query: 316 LHLTYSRHTDLNVKAFSDKSRDYTV---------LDPSLHAAQAPAWQTTQAATMYSGSM 366
LH++YSRHTDL++K +++SRDYT+ PS+ Q P T Y+G+
Sbjct: 410 LHISYSRHTDLSIKVNNERSRDYTIPNVPPPVVNAQPSI-LGQHPVPMTGPPPQHYNGA- 583
Query: 367 GQMPSWDPNQQEVTQSYLSAP-GTFP--SGQAAPPFPGYSPAAVPPAGASPHSHMP 419
+ P ++ S+ A GT P S Q P Y+P+A+ P +P P
Sbjct: 584 ----QYAPVTEQTLMSHSQAGWGTGPPQSMQQMHNHP-YTPSAMVPPQTTPGMQFP 736
Score = 102 bits (254), Expect = 4e-22
Identities = 56/108 (51%), Positives = 72/108 (65%)
Frame = +2
Query: 111 NVLLVTIEGVEAGDVSIDVIHLVFSAFGFVHKLATFEKTAGFQALIQYTDAETAASAKDS 170
NVLL +IE ++ V++DV+H+VFSAFG V K+A F+K G QALIQY D +TA AK++
Sbjct: 194 NVLLASIENMQY-TVTLDVLHMVFSAFGPVQKIAMFDKNGGLQALIQYPDIQTAVVAKET 370
Query: 171 LDGRSIPRYLLPEHVGACNLRISYSAHRDLNIKFQSNRSRDYTNPMLP 218
L+G I G C L ISYS H DL+IK + RSRDYT P +P
Sbjct: 371 LEGHCI------YDGGFCKLHISYSRHTDLSIKVNNERSRDYTIPNVP 496
Score = 38.1 bits (87), Expect = 0.008
Identities = 17/33 (51%), Positives = 21/33 (63%)
Frame = +1
Query: 180 LLPEHVGACNLRISYSAHRDLNIKFQSNRSRDY 212
L E++G C L+I YS H D F+ NRSRDY
Sbjct: 1 LFLENIGPCGLKIKYSGHSD*VSSFKVNRSRDY 99
>BU081841 homologue to GP|9757999|dbj polypyrimidine tract-binding RNA
transport protein-like {Arabidopsis thaliana}, partial
(26%)
Length = 425
Score = 174 bits (442), Expect = 6e-44
Identities = 81/113 (71%), Positives = 97/113 (85%)
Frame = +3
Query: 1 MSSSKQQQQFRCTQIPSKVLHFRNLPWECSDEELADLCSPFGKVMSTKCNVGVNQNQAFV 60
M+S QQQFR TQ PSKVLH RNLPWEC++EEL +L PFGKV++TKCNVG N+NQAF+
Sbjct: 87 MASVSSQQQFRYTQPPSKVLHLRNLPWECTEEELIELGKPFGKVVNTKCNVGANRNQAFI 266
Query: 61 QFAELNQAISMVSYYASSSEPAQVRGKTVYIQYSNRHHIVNSNSPGDIPGNVL 113
+FA+LNQAI+M+SYYASSSEPAQVRGKTVY+QYSNR IVN+ + D+ GNVL
Sbjct: 267 EFADLNQAIAMISYYASSSEPAQVRGKTVYLQYSNRQEIVNNKTAADVAGNVL 425
>TC205238 similar to UP|Q6ICX4 (Q6ICX4) At1g43190, partial (98%)
Length = 1560
Score = 155 bits (391), Expect = 5e-38
Identities = 106/344 (30%), Positives = 176/344 (50%), Gaps = 33/344 (9%)
Frame = +1
Query: 16 PSKVLHFRNLPWECSDEELADLCSPFGKVMSTKCNVGVNQNQAFVQFAELNQAISMVSYY 75
PSKV+H RN+ E S+ +L L PFG + TK + +NQA +Q ++ A++ + +Y
Sbjct: 76 PSKVIHVRNVGHEISENDLLQLFQPFGVI--TKLVMLRAKNQALLQMQDIPSAVNALQFY 249
Query: 76 ASSSEPAQVRGKTVYIQYSNRHHIV---NSNSPGDIPGNVLLVTIEGVEAGDVSIDVIHL 132
A+ +RG+ VY+Q+S+ + + + D P +LLVT+ + ++ DV+H
Sbjct: 250 ANVQ--PSIRGRNVYVQFSSHQELTTMDQNQAREDEPNRILLVTVHHM-LYPITADVLHQ 420
Query: 133 VFSAFGFVHKLATFEKTAGFQALIQYTDAETAASAKDSLDGRSIPRYLLPEHVGACNLRI 192
VFS GFV K+ TF+K+AGFQALIQY ++A +A+ +L GR+I + G C L I
Sbjct: 421 VFSPHGFVEKIVTFQKSAGFQALIQYQSRQSAVTARSTLQGRNI-------YDGCCQLDI 579
Query: 193 SYSAHRDLNIKFQSNRSRDYTNPMLP------------------------------VNQA 222
+S +L + + ++RSRD+TNP LP N A
Sbjct: 580 QFSNLDELQVNYNNDRSRDFTNPNLPTEQKGRSSQPGYGDAAGMHSGARAGGFSQMANAA 759
Query: 223 AIDSALQPAIGPDGKRKEHKSNVLLATIENMQYAVPLDVLHSVFSAFGFVQKVAMFDKNG 282
AI +A + P + VL+A + + + D + ++FS +G + ++ + +N
Sbjct: 760 AIAAAFGGGLPPGITGTNERCTVLVANLNPDR--IDEDKMFNLFSIYGNIVRIKLL-RNK 930
Query: 283 HTHALIQYPDLTIAAAAKETLEGHCIYDGGYCKLHLTYSRHTDL 326
HALIQ D A A L+G ++ +L + YS+H ++
Sbjct: 931 PDHALIQMGDGFQAELAVHFLKGAMLFGK---RLEVNYSKHANI 1053
Score = 91.3 bits (225), Expect = 8e-19
Identities = 77/254 (30%), Positives = 118/254 (46%), Gaps = 12/254 (4%)
Frame = +1
Query: 124 DVSIDVIHLVFSAFGFVHKLATFEKTAGFQALIQYTDAETAASA-------KDSLDGRSI 176
++S + + +F FG + KL A QAL+Q D +A +A + S+ GR
Sbjct: 112 EISENDLLQLFQPFGVITKLVMLR--AKNQALLQMQDIPSAVNALQFYANVQPSIRGR-- 279
Query: 177 PRYLLPEHVGACNLRISYSAHRDLNIKFQSNRSRDYTNPMLPVNQAAIDSALQPAIGPDG 236
N+ + +S+H++L Q NQA
Sbjct: 280 ------------NVYVQFSSHQELTTMDQ--------------NQA-------------- 339
Query: 237 KRKEHKSNVLLATIENMQYAVPLDVLHSVFSAFGFVQKVAMFDKNGHTHALIQYPDLTIA 296
R++ + +LL T+ +M Y + DVLH VFS GFV+K+ F K+ ALIQY A
Sbjct: 340 -REDEPNRILLVTVHHMLYPITADVLHQVFSPHGFVEKIVTFQKSAGFQALIQYQSRQSA 516
Query: 297 AAAKETLEGHCIYDGGYCKLHLTYSRHTDLNVKAFSDKSRDYTVLDPSLHAAQ-----AP 351
A+ TL+G IYD G C+L + +S +L V +D+SRD+T +P+L Q P
Sbjct: 517 VTARSTLQGRNIYD-GCCQLDIQFSNLDELQVNYNNDRSRDFT--NPNLPTEQKGRSSQP 687
Query: 352 AWQTTQAATMYSGS 365
+ AA M+SG+
Sbjct: 688 GY--GDAAGMHSGA 723
>AW757383 similar to GP|9757999|dbj polypyrimidine tract-binding RNA
transport protein-like {Arabidopsis thaliana}, partial
(25%)
Length = 337
Score = 141 bits (355), Expect = 7e-34
Identities = 66/112 (58%), Positives = 86/112 (75%)
Frame = +2
Query: 2 SSSKQQQQFRCTQIPSKVLHFRNLPWECSDEELADLCSPFGKVMSTKCNVGVNQNQAFVQ 61
+S QQQ R TQ PS VLH RNLPWE ++EEL +L PFGKV++TKCNVG N+NQ+ ++
Sbjct: 2 ASESSQQQIRYTQPPSNVLHLRNLPWEWTEEELIELGKPFGKVVNTKCNVGANRNQSIIE 181
Query: 62 FAELNQAISMVSYYASSSEPAQVRGKTVYIQYSNRHHIVNSNSPGDIPGNVL 113
FA++ AI+M+SYYASS+EPAQV+G TV +QYSNR I N + D+ G+VL
Sbjct: 182 FADVTHAIAMISYYASSTEPAQVQGTTVCLQYSNRQVIRNHKTAADVAGDVL 337
>AW596276
Length = 202
Score = 118 bits (295), Expect = 6e-27
Identities = 53/66 (80%), Positives = 59/66 (89%)
Frame = +2
Query: 275 VAMFDKNGHTHALIQYPDLTIAAAAKETLEGHCIYDGGYCKLHLTYSRHTDLNVKAFSDK 334
+A+F+KNG T ALIQYPD+ A+AA E LEGHCIYDGGYCKLHL+YSRHTDLNVKA SDK
Sbjct: 2 IAIFEKNGQTQALIQYPDIITASAATEALEGHCIYDGGYCKLHLSYSRHTDLNVKALSDK 181
Query: 335 SRDYTV 340
SRDYTV
Sbjct: 182 SRDYTV 199
Score = 63.9 bits (154), Expect = 1e-10
Identities = 35/73 (47%), Positives = 45/73 (60%)
Frame = +2
Query: 143 LATFEKTAGFQALIQYTDAETAASAKDSLDGRSIPRYLLPEHVGACNLRISYSAHRDLNI 202
+A FEK QALIQY D TA++A ++L+G I G C L +SYS H DLN+
Sbjct: 2 IAIFEKNGQTQALIQYPDIITASAATEALEGHCI------YDGGYCKLHLSYSRHTDLNV 163
Query: 203 KFQSNRSRDYTNP 215
K S++SRDYT P
Sbjct: 164 KALSDKSRDYTVP 202
>TC208609 similar to UP|O82472 (O82472) Polypyrimidine tract-binding protein
homolog, partial (5%)
Length = 853
Score = 111 bits (278), Expect = 6e-25
Identities = 61/103 (59%), Positives = 67/103 (64%), Gaps = 12/103 (11%)
Frame = +2
Query: 330 AFSDKSRDYTVLDPSLHAAQAPA--WQTTQAATMYSGSM---------GQMPSWDPNQQE 378
AFSDKSRDYTV DPSL AAQ PA WQ QAA MY GS GQ+P+WDPN Q
Sbjct: 185 AFSDKSRDYTVPDPSLLAAQGPATAWQNPQAAPMYPGSAPAYHTQVPGGQVPAWDPNLQA 364
Query: 379 VTQSYLSAPGTF-PSGQAAPPFPGYSPAAVPPAGASPHSHMPP 420
V SY+SAPGTF AAPP P Y+PAA PA +SPH+ P
Sbjct: 365 VRPSYVSAPGTFHVQSGAAPPMPAYAPAAAMPAASSPHAQSSP 493
>TC227164 similar to UP|Q9FGL9 (Q9FGL9) Polypyrimidine tract-binding RNA
transport protein-like, partial (11%)
Length = 409
Score = 80.1 bits (196), Expect = 2e-15
Identities = 48/131 (36%), Positives = 67/131 (50%), Gaps = 10/131 (7%)
Frame = +2
Query: 299 AKETLEGHCIYDGGYCKLHLTYSRHTDLNVKAFSDKSRDYTV---------LDPSLHAAQ 349
AKE LEGHCIYDGG+CKLH++YSRH+DL++K +D+SRDYT+ PS+ Q
Sbjct: 2 AKEALEGHCIYDGGFCKLHISYSRHSDLSIKVNNDRSRDYTIPNVPPPVVNAQPSI-LGQ 178
Query: 350 APAWQTTQAATMYSGSMGQMPSWDPNQQEVTQSYLSAPGTFPSGQAAPPFPG-YSPAAVP 408
P T Y+G+ +Q + + GT PS Y+P+A+
Sbjct: 179 HPVPMTGPPPQQYNGAQYA----SVTEQTLMPQSQAGWGTAPSQSMLQMHNNPYTPSAMV 346
Query: 409 PAGASPHSHMP 419
P +P P
Sbjct: 347 PPQTAPGMQFP 379
Score = 46.6 bits (109), Expect = 2e-05
Identities = 25/52 (48%), Positives = 32/52 (61%)
Frame = +2
Query: 167 AKDSLDGRSIPRYLLPEHVGACNLRISYSAHRDLNIKFQSNRSRDYTNPMLP 218
AK++L+G I G C L ISYS H DL+IK ++RSRDYT P +P
Sbjct: 2 AKEALEGHCI------YDGGFCKLHISYSRHSDLSIKVNNDRSRDYTIPNVP 139
>CD394720
Length = 560
Score = 68.2 bits (165), Expect = 7e-12
Identities = 31/55 (56%), Positives = 37/55 (66%), Gaps = 1/55 (1%)
Frame = -1
Query: 367 GQMPSWDPNQQEVTQSYLSAPGTFP-SGQAAPPFPGYSPAAVPPAGASPHSHMPP 420
GQ+PSWDP+ Q V SY SAPGTFP AAPP P Y+PA+ P +SPH+ P
Sbjct: 536 GQVPSWDPSLQAVRPSYASAPGTFPVQTGAAPPMPSYAPASAMPVASSPHAQSSP 372
>TC231826 similar to UP|Q6ICX4 (Q6ICX4) At1g43190, partial (20%)
Length = 508
Score = 56.2 bits (134), Expect = 3e-08
Identities = 29/84 (34%), Positives = 50/84 (59%)
Frame = +1
Query: 16 PSKVLHFRNLPWECSDEELADLCSPFGKVMSTKCNVGVNQNQAFVQFAELNQAISMVSYY 75
PSKV+H RN+ E S+ +L L PFG + TK + +NQA +Q ++ A++ + +Y
Sbjct: 253 PSKVIHVRNVGHEISENDLLQLFQPFGVI--TKLVMLRAKNQALIQMQDVPSAVNALQFY 426
Query: 76 ASSSEPAQVRGKTVYIQYSNRHHI 99
A+ +RG+ VY+Q+S+ +
Sbjct: 427 ANVQ--PSIRGRNVYVQFSSHXEL 492
>TC205242 similar to UP|Q6ICX4 (Q6ICX4) At1g43190, partial (69%)
Length = 1451
Score = 51.6 bits (122), Expect = 7e-07
Identities = 26/65 (40%), Positives = 41/65 (63%)
Frame = +3
Query: 146 FEKTAGFQALIQYTDAETAASAKDSLDGRSIPRYLLPEHVGACNLRISYSAHRDLNIKFQ 205
F+K+AGFQALIQY ++A +A+ +L GR+I + G C L I +S +L + +
Sbjct: 3 FQKSAGFQALIQYQSRQSAVAARSTLQGRNI-------YDGCCQLDIQFSNLDELQVNYN 161
Query: 206 SNRSR 210
++RSR
Sbjct: 162 NDRSR 176
Score = 44.3 bits (103), Expect = 1e-04
Identities = 25/59 (42%), Positives = 34/59 (57%)
Frame = +3
Query: 278 FDKNGHTHALIQYPDLTIAAAAKETLEGHCIYDGGYCKLHLTYSRHTDLNVKAFSDKSR 336
F K+ ALIQY A AA+ TL+G IYD G C+L + +S +L V +D+SR
Sbjct: 3 FQKSAGFQALIQYQSRQSAVAARSTLQGRNIYD-GCCQLDIQFSNLDELQVNYNNDRSR 176
>TC224751 similar to UP|Q6V8K5 (Q6V8K5) GPRP, partial (72%)
Length = 851
Score = 44.3 bits (103), Expect = 1e-04
Identities = 22/56 (39%), Positives = 28/56 (49%), Gaps = 2/56 (3%)
Frame = +1
Query: 382 SYLSAPGTFPSGQAAPPFPGYSPAAVPPAGASPHSHMPPSSFAGA--FPGSQPHYG 435
+Y PG +P Q PP GY PA PP P + PP+ + G+ PGS H G
Sbjct: 367 AYPPPPGAYPPHQGYPPQHGYPPAGYPPHQGYPPAGYPPAGYPGSSHAPGSHGHGG 534
>TC224716 similar to GB|AAP21369.1|30102902|BT006561 At4g19200 {Arabidopsis
thaliana;} , partial (67%)
Length = 883
Score = 40.0 bits (92), Expect = 0.002
Identities = 22/51 (43%), Positives = 27/51 (52%), Gaps = 2/51 (3%)
Frame = +1
Query: 382 SYLSAPGTFPSGQAAPPFPGYSPA-AVPPAGASPHSHMPPSSFAGA-FPGS 430
+Y PG +P Q PP GY P PPAG PH PP+ + A +PGS
Sbjct: 277 AYPPPPGAYPPQQGYPP-AGYPPQQGYPPAGYPPHQGYPPAGYPPAGYPGS 426
Score = 36.2 bits (82), Expect = 0.031
Identities = 34/122 (27%), Positives = 47/122 (37%), Gaps = 7/122 (5%)
Frame = +1
Query: 307 CIYDGGYCKLHLTYSRHTDLNVKAF------SDKSRDYTVLDPSLHAAQAPAWQTTQAAT 360
C+++ C + +YS +++ V F DK D + H A A
Sbjct: 88 CVHE*EPCSITTSYSANSN-RVLVF*KMGGGKDKHHDESDKGIFSHLAHGVAGAAHGGHG 264
Query: 361 MYSGSMGQMP-SWDPNQQEVTQSYLSAPGTFPSGQAAPPFPGYSPAAVPPAGASPHSHMP 419
G+ P ++ P Q Y G P+G PP GY PA PPAG SH P
Sbjct: 265 YPPGAYPPPPGAYPPQQGYPPAGYPPQQGYPPAGY--PPHQGYPPAGYPPAGYPGSSHAP 438
Query: 420 PS 421
S
Sbjct: 439 GS 444
Score = 29.6 bits (65), Expect = 2.9
Identities = 17/48 (35%), Positives = 20/48 (41%), Gaps = 1/48 (2%)
Frame = +1
Query: 390 FPSGQAAPPFPGYSPA-AVPPAGASPHSHMPPSSFAGAFPGSQPHYGW 436
+P G PP Y P PPAG P PP+ G PH G+
Sbjct: 265 YPPGAYPPPPGAYPPQQGYPPAGYPPQQGYPPA-------GYPPHQGY 387
>TC217837 similar to PIR|S65780|S65780 glycine/proline-rich protein GPRP -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(40%)
Length = 891
Score = 39.3 bits (90), Expect = 0.004
Identities = 23/67 (34%), Positives = 31/67 (45%), Gaps = 5/67 (7%)
Frame = +2
Query: 375 NQQEVTQSYLSAPGTF-----PSGQAAPPFPGYSPAAVPPAGASPHSHMPPSSFAGAFPG 429
N+++ T S G F PS PP GY P+ PP G P + PP ++ PG
Sbjct: 191 NRKDNTDDESSERGIFSHLGYPSAPPYPPPHGYPPSGYPPPGGYPPTAYPPPAYPP--PG 364
Query: 430 SQPHYGW 436
PH G+
Sbjct: 365 VYPHSGY 385
Score = 36.2 bits (82), Expect = 0.031
Identities = 26/70 (37%), Positives = 30/70 (42%), Gaps = 16/70 (22%)
Frame = +2
Query: 383 YLSAP------GTFPSGQAAP---PFPGYSPAAVPPAGASPHSHMPPSSFAGAF------ 427
Y SAP G PSG P P Y P A PP G PHS PS + A+
Sbjct: 251 YPSAPPYPPPHGYPPSGYPPPGGYPPTAYPPPAYPPPGVYPHSGYYPSEYPPAYPPPGGY 430
Query: 428 -PGSQPHYGW 436
P + PH G+
Sbjct: 431 PPTTYPHSGY 460
>BF425720
Length = 178
Score = 39.3 bits (90), Expect = 0.004
Identities = 20/49 (40%), Positives = 27/49 (54%)
Frame = +1
Query: 188 CNLRISYSAHRDLNIKFQSNRSRDYTNPMLPVNQAAIDSALQPAIGPDG 236
C L I +S +L + + ++RSRD+TNP LP Q S QP G G
Sbjct: 1 CQLDIQFSNLDELQVNYNNDRSRDFTNPNLPTEQKGRPS--QPGYGDAG 141
>TC227129 similar to UP|Q6V8K5 (Q6V8K5) GPRP, partial (62%)
Length = 1117
Score = 37.0 bits (84), Expect = 0.018
Identities = 22/55 (40%), Positives = 26/55 (47%), Gaps = 3/55 (5%)
Frame = +1
Query: 382 SYLSAPGTFPSGQAAPPFPGYSP-AAVPPAGASPHSHMPPSSFAGAFP--GSQPH 433
+Y PG +P Q PP GY P PPAG P + P AG +P G PH
Sbjct: 259 AYPPPPGAYPPYQGYPPAHGYPPQGGYPPAGYPPAGYPPA---AGGYPPAGYPPH 414
Score = 30.0 bits (66), Expect = 2.2
Identities = 23/60 (38%), Positives = 25/60 (41%), Gaps = 5/60 (8%)
Frame = +1
Query: 381 QSYLSAPGTFPSGQAAP---PFPGYSPAA--VPPAGASPHSHMPPSSFAGAFPGSQPHYG 435
Q Y A G P G P P GY PAA PPAG PH +PGS +G
Sbjct: 295 QGYPPAHGYPPQGGYPPAGYPPAGYPPAAGGYPPAGYPPH----------GYPGSSAPHG 444
Score = 28.5 bits (62), Expect = 6.4
Identities = 17/45 (37%), Positives = 20/45 (43%), Gaps = 5/45 (11%)
Frame = +1
Query: 387 PGTFPSGQAAPPFPGYSPAAVPPAG----ASPH-SHMPPSSFAGA 426
P +P P GY PA PP G ++PH SH P GA
Sbjct: 343 PAGYPPAGYPPAAGGYPPAGYPPHGYPGSSAPHGSHAPGHGGMGA 477
>CD416342 similar to SP|P42698|D111_ DNA-damage-repair/toleration protein
DRT111 chloroplast precursor. [Mouse-ear cress],
partial (24%)
Length = 624
Score = 37.0 bits (84), Expect = 0.018
Identities = 28/91 (30%), Positives = 37/91 (39%)
Frame = -1
Query: 339 TVLDPSLHAAQAPAWQTTQAATMYSGSMGQMPSWDPNQQEVTQSYLSAPGTFPSGQAAPP 398
T P AP W ++A+ S PS+ P S P + S PP
Sbjct: 357 TTSPPPTSGPPAPRWLPPRSASPPPSSRPPKPSYGPTPSP------SPPPSPSSPPPPPP 196
Query: 399 FPGYSPAAVPPAGASPHSHMPPSSFAGAFPG 429
FP +P + PP+ AS PPSS + PG
Sbjct: 195 FPRKTPPSSPPSLASS----PPSSRSTTRPG 115
Score = 32.7 bits (73), Expect = 0.34
Identities = 16/42 (38%), Positives = 17/42 (40%)
Frame = -1
Query: 391 PSGQAAPPFPGYSPAAVPPAGASPHSHMPPSSFAGAFPGSQP 432
P + PP P Y P P SP S PP F P S P
Sbjct: 291 PPPSSRPPKPSYGPTPSPSPPPSPSSPPPPPPFPRKTPPSSP 166
>TC203569 similar to GB|AAO63422.1|28950997|BT005358 At1g03457 {Arabidopsis
thaliana;} , partial (23%)
Length = 553
Score = 36.6 bits (83), Expect = 0.024
Identities = 27/81 (33%), Positives = 39/81 (47%), Gaps = 4/81 (4%)
Frame = +1
Query: 16 PSKVLHFRNLPWECSDEELADLCSPFGKVMSTKCNV----GVNQNQAFVQFAELNQAISM 71
P L ++P E DEELA PFG+V+S K V GV++ FV + A S
Sbjct: 226 PGANLFIYHIPQEYGDEELATTFQPFGRVLSAKIFVDKVTGVSKCFGFVSYDTPEAAQSA 405
Query: 72 VSYYASSSEPAQVRGKTVYIQ 92
+ S+ Q+ GK + +Q
Sbjct: 406 I----STMNGCQLGGKKLKVQ 456
>TC224715 homologue to UP|Q39882 (Q39882) Nodulin-26, partial (98%)
Length = 1270
Score = 35.0 bits (79), Expect = 0.069
Identities = 30/91 (32%), Positives = 39/91 (41%), Gaps = 2/91 (2%)
Frame = +2
Query: 344 SLHAAQAPAWQTTQAATMYSGSM-GQMPSWDPNQQEVTQSYLSAPGTFPSGQAAPPFPGY 402
S AQAPA TT + T + P P + SAP T P+ + PP P
Sbjct: 599 SCSPAQAPASPTTSSPTTALPPLPASSPPPSPMHSPSSSPSPSAP-TSPAATSTPPSPSA 775
Query: 403 -SPAAVPPAGASPHSHMPPSSFAGAFPGSQP 432
S AA P+ A+ + P SS + P S P
Sbjct: 776 PSSAATSPSSAASSTSSPSSSAPSSPPSSWP 868
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.131 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,083,123
Number of Sequences: 63676
Number of extensions: 295848
Number of successful extensions: 3301
Number of sequences better than 10.0: 199
Number of HSP's better than 10.0 without gapping: 2592
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3076
length of query: 449
length of database: 12,639,632
effective HSP length: 100
effective length of query: 349
effective length of database: 6,272,032
effective search space: 2188939168
effective search space used: 2188939168
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148485.4


